Copernicus, Kepler, Galileo, Newton—these are familiar names. During a 150-year span in the sixteenth and seventeenth centuries, they led the scientific revolution that placed the sun, rather than the earth, at the center of things astronomical. But have you ever heard of Carl Woese? He set in motion a scientific revolution in biology that, in its repudiation of anthropocentric views of life, is proving no less profound.
In 1977, Woese (pronounced “woes”), a professor at the University of Illinois at Urbana-Champaign, drew a terrestrial family tree that showed the genetic relatedness of all living things on this planet. Using modern tools of molecular biology, he sampled the known single-celled, microscopic organisms we call microbes, and also the denizens of the human-scale world with which we are familiar: the flowers, trees, and shrubs; the animals; and the fungi. His map of all this new information revealed that taxonomists of ages past had been looking at the world through the wrong end of a telescope. The formerly great “kingdoms” of Plantae, Animalia, and Fungi almost disappeared, shrinking to fit on a small, trifurcating branch of his tree. In their place were three vast “domains”: Bacteria (single-celled microorganisms that lack a distinct nucleus and organelles); Archaea, or Archaebacteria (similar in appearance and simplicity to bacteria, but with notably different molecular organization); and Eukarya (all organisms whose cells have a distinct nucleus—or, simply put, everything else). Life on Earth, Woese’s model showed, is overwhelmingly microbial. In fact, the extent of microbial diversity is so great that scientists have difficulties estimating its actual size. Some estimates place the number of microbial species in the range of billions, exceeding the number of species of “large” organisms by several orders of magnitude.

The anthropocentric five-kingdom system classified all unicellular organisms lacking nuclei (archaea and bacteria) as Monera. The nucleated eukaryotes comprising plants, animals, and fungi were thought to represent the bulk of biological diversity. All other nucleated eukaryotes were grouped in a grab-bag classification known as Protista.

The modern “tree of life,” based on genetic analysis, shows that the bulk of Earth’s biodiversity resides among the Archaea, Bacteria, and that portion of the Eukarya that does not include plants, animals, and fungi.
In light of this new understanding of life, scientists with advanced research tools are focusing anew on microbes, which, following the great discoveries of penicillin and other antibiotics in the mid twentieth century, had largely been consigned to the confines of pharmaceutical research.
“Our planet has been shaped by an invisible world,” says Roberto Kolter, a professor of microbiology and molecular genetics at Harvard Medical School (HMS). He and Jeffrey professor of biology Colleen Cavanaugh of the Faculty of Arts and Sciences (FAS) together co-direct Harvard’s Microbial Sciences Initiative, which serves as a focal point for researchers in the field from all over the University. “Microbes mediate all the important element cycles on Earth, and have played a defining role in the development of the planet,” says Kolter. They form clouds, break down rocks, deposit minerals, fertilize plants, condition soils, and clean up toxic waste. Among their ranks, explains Cavanaugh, are the photosynthetic “primary producers” that use sunlight, water, and carbon dioxide to form the broad base of the food chain, and together with plants make up the earth’s largest source of biomass. The earliest life on our planet was entirely microbial, and if life exists on other planets, it is surely microbial there as well.


In terms of gene content, humans and potatoes are more closely related than these two bacteria are to each other—one measure of bacterial diversity. On the left, Vibrio cholerae; on the right, Mycobacterium tuberculosis.

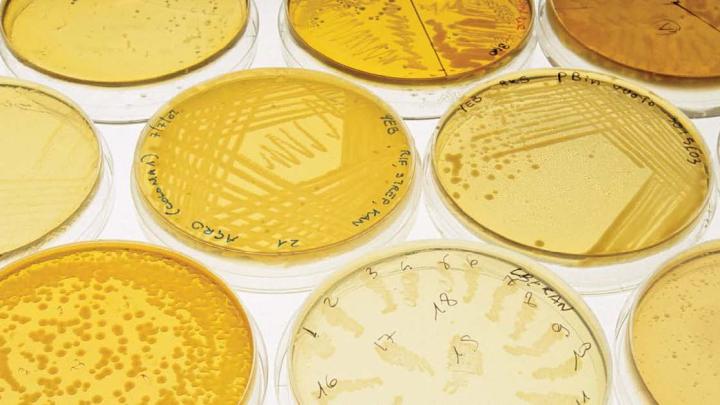
From the billions of bacteria in a soil sample, these few cells landed on a nutrient medium where they could grow and form colonies. “There is so much beauty everywhere we look,” says microbiologist Roberto Kolter.

A strain of Streptomyces that produces several antibiotics widely used in medicine and agriculture. When starved, these bacteria take on a “hairy” appearance as they initiate growth of aerial structures where protective spores develop. The pigmented areas consist of small molecules, often with antibiotic properties.

In the presence of the antibiotic streptomycin, colorful antibiotic-resistant mutants of Streptomyces coelicolor spring up and form colonies.
In the realm of human health, microbes help us digest food and produce vitamins, protect us against infection, and are the main source of antibiotic medicines. The human cells in your body number 10 trillion, but that pales by comparison to the estimated 100 trillion microbial cells that live in and on you. “Without them, you would be in trouble,” Kolter says: animals experience abnormal growth and become sick if deprived of their microflora during development. Although a few microbes are known to cause disease, the precise role played by the vast majority is essentially unknown.
The same could be said for microbes around the planet. There are a billion of them in a gram of soil, and a billion per liter of seawater, but we know neither what they are nor what they do.
In the poetic conclusion to his 1994 autobiography, Naturalist, the great sociobiologist and Pellegrino University Professor emeritus E.O. Wilson mused on what he would do, “[i]f I could do it all over again and relive my vision in the twenty-first century. I would be a microbial ecologist...,” he wrote. “Into that world I would go with the aid of modern microscopy and molecular analysis. I would cut my way through clonal forests sprawled across grains of sand, travel in an imagined submarine through drops of water proportionately the size of lakes, and track predators and prey in order to discover new life ways and alien food webs. All this, and I need venture no farther than ten paces outside my laboratory building. The jaguars, ants, and orchids would still occupy distant forests in all their splendor, but now they would be joined by an even stranger and vastly more complex living world virtually without end.”
The Limits of Cultures
As a practical matter, the Microbial Sciences Initiative (MSI) began in 2002 as a grass-roots effort among faculty members who recognized the unexplored ecology and potential of these organisms and wanted to share information about microbial research across diverse disciplines: biology, medicine, chemistry, engineering, geology, astronomy, and evolutionary and planetary history. The group held informal “chalk-talks” weekly, and in 2004 staged a day-long symposium with speakers from around the world. When President Lawrence H. Summers issued a call that year for initiatives that would bring people together from across the science and engineering disciplines, MSI was a perfect candidate, says Cabot professor of biology Richard Losick, a member of its steering committee. “I think there are few disciplines that lend themselves better to cross-disciplinary approaches,” he says, “and few subjects that have implications across a wider spectrum of sciences than is true for microbiology.” As a result, in 2006 MSI received four years of support, totaling $2.7 million, from the provost’s office.
“It kills me that people think only that bacteria are disease-causing,” says Cavanaugh, who studies the chemosynthetic symbiotic bacteria that make life possible for giant clams and tubeworms dwelling near deep ocean hydrothermal vents. Far from sunlight, they operate by mechanisms both similar to and much different from the photosynthetic organisms we see every day. “Although intracellular, these bacteria are helpful to their animal hosts,” she adds. “Like chloroplasts in plants [which evolved from symbiotic photosynthetic bacteria], the chemosynthetic symbionts turn carbon dioxide into sugars and proteins, feed- ing their hosts internally.”


Not much can be gleaned about the differences between these two microorganisms just by looking at them, but genetic analysis tells us that they are not even in the same domain. Left: Soil-dwelling Bacillus anthracis is classified as a bacterium. Right: Heat-loving Thermoproteus tenax is an archaean that lives in hot springs.
But most people do associate microbes with disease. “Antibacterials” have been incorporated into all kinds of consumer products: soaps, sponges, toilet paper, towels, and cutting boards—even clothing. Kolter traces the origins of this “ludicrous” antimicrobial “scorched-earth policy” to the time of Louis Pasteur, who formulated germ theory, and Robert Koch, who developed methods for culturing bacteria. “Medical microbiology for almost 150 years has been driven by the idea that germs are the causative agents of disease. And there is no doubt that Koch and Pasteur were right, that Mycobacterium tuberculosis causes tuberculosis and Vibrio cholerae causes cholera,” says Kolter. But microbes have also led to most of our antibiotics, a development that Kolter calls “the most important advance in medical history.”
Scientists had known that there are more microbes in an ounce of soil than humans alive on Earth, but that was just a measure of abundance. Pace’s discovery demonstrated something new, a previously unfathomed repository of biodiversity. Scientists began sequencing DNA from all sorts of environments. After looking at human gut microflora, they learned that each individual has his or her own characteristic set of a thousand species. “These represent three million genes that you carry,” points out Kolter, “as compared to the estimated 18,000 genes of the human genome. So you are living and exchanging [metabolites] constantly with a diverse pool of some three million genes.” Microbiologists continue to find new taxonomic divisions of microbes far faster than they can figure out how to culture them.

Their cell membranes outlined in red, individual Bacillus subtilis bacteria contain the biosynthetic machinery—shown as green dots—for making bacillaene, a newly discovered small molecule.

Colonies formed by the wild strain of Bacillus subtilis generate complex structures, as seen at left, while the domesticated strain—presumably selected for uniformity by generations of researchers who have used it in laboratory experiments for many decades—has lost nearly all colony architecture.
The formerly limited view of the microbial world arose from what has turned out to be an inherently constrained approach to the study of bacteria: the practice of culturing them. For more than a hundred years, scientists had been mystified by what was called the “plate count paradox.” Whenever they tried to grow a sample of bacteria from the environment on a nutrient medium in a petri dish (an agar plate), only a few microorganisms grew and multiplied to form colonies, when there should have been at a minimum thousands of such colonies (based on the number of different species discernible just by looking through a microscope). Various explanations were offered—that 99.9 percent of the bacteria in the sample were dead, or that they must all be the same bacterium because they looked similar.
“But then in 1990,” says Kolter, “scientists showed that the DNA complexity in a typical soil sample meant that there had to be thousands of times more diversity than was being plated.” Norman Pace, a professor at the University of Colorado, began to wonder if scientists simply lacked the basic knowledge needed to grow most of the bacteria on the planet—if perhaps we were so ignorant of these bacteria that we could not culture them. He hit on the idea that he could instead analyze a sample of soil or water for its DNA content in order to ascertain how many species it contained. “Pace went to Yellowstone National Park, to some of its famous hot springs, where the water was nearly boiling, and collected a sample of sediment,” Kolter explains. “He extracted the DNA, cloned it, and put it into a little bacterium that he knew how to grow.” Then he sequenced the genes. “In that one little gram of sediment,” Kolter notes, “Pace discovered more diversity than we ever knew existed before, when using our traditional, century-old techniques for cultivating bacteria.”
The world of animals—from elephants to ants—is divided into 13 phyla (vertebrates are one phylum, insects another). In the microbial world, their equivalents are called, for the time being, “deep-rooting branches.” In 1987, 13 of these big divisions were known in the bacterial domain: Woese sampled these to create his tree of life. But by 1997, there were almost three times as many: 36 in all. “Twelve of them we had never cultivated, and the others we began to learn how to cultivate,” Kolter explains. “By 2003, there were 53 divisions, but the more we discovered, the more we found representative microbes that we could not cultivate. We learned how to cultivate microbes from only two of these in six years and by 2004 we had found 80 such divisions from which we couldn’t cultivate even a single representative.” Each of these deep-branching divisions is thought to represent millions, if not hundreds of millions, of species. “That means there are lots of genes out there, and we have no clue what they are doing,” Kolter says. “So when you think about biodiversity, and the extent of diversity on the planet, you really get a sense of how little we know about this undiscovered world. We are at the stage of discovery where, everywhere we look, we see new species.
“The reason we are so excited about preserving diversity,” he continues, “is because that is how we preserve the richness of the planet.” Where does that richness lie? Traditionally, taxonomists classified life according to morphology (by appearance, form, or pattern): one finch’s beak resembles that of another, but is nothing like that of hummingbird. Bats fly, but are more like mice with wings than like birds. Microbes, on the other hand, often appear similar to one another to the human eye. Plants, animals, and fungi may seem wildly diverse from a morphological perspective, but in fact, Kolter explains, there is “far less genetic difference between [a human being] and a potato” than there is between, say, “the bacterium that causes tuberculosis and the one that causes cholera. So as magnificent as is the diversity of the tropical rainforest, it pales by comparison to the microbial world.”
Microbial Medicines
In the interests of human health, researchers have begun to hunt among all this newfound microbial diversity for new antibiotics. Says Richard Losick, “I have many wonderful colleagues in the chemistry department, but without question the champion synthetic organic chemists on the planet are the microbes.”
“The majority of our antibiotics and many anticancer compounds come from soil-dwelling bacteria,” notes Jon Clardy, a professor of biological chemistry and molecular pharmacology at HMS. Unfortunately, he says, due to the spread of antibiotic resistance, “we are running out of effective antibiotics.” For economic reasons, pharmaceutical companies are not investing as much as they used to in the development of new ones, which has left physicians looking for an antibiotic of last resort to be held in reserve for the one patient once a year who has a resistant strain of tuberculosis or pneumonia, explains Kolter. “That drug would be the billion-dollar blockbuster that nobody buys, because doctors shouldn’t be prescribing it widely. Nobody in the corporate world will develop it.” He believes that “there may be a role for the University in research that is not going to be as profitable for corporations to pursue: the development of targeted, ecologically sound antibiotics.”

Two colonies of Bacillus subtilis growing on top of another bacterium, Streptomyces coelicolor. The Bacillus colonies “talk” to the Streptomyces using small molecules. The strain of Bacillus subtilis at left makes compounds for turning production of a small pigmented molecule in Streptomyces on and off. But in a mutant strain of Bacillus that cannot produce the “off switch,” Streptomyces produces the pigmented red ring. This led to the discovery of the molecule that acts as the off switch: bacillaene. Below, the molecule’s chemical structure.

Whole-genome studies of microbes suggest that only a small fraction of the natural products that come from even well-known bacteria have been discovered. A prime example is Streptomyces avermitilis, the bacterium from which researchers derived a drug called Ivermectin. “Ivermectin,” says Clardy, “is grown on the ton scale because it is used against river blindness in Africa, for treating almond trees in California, and for getting rid of all kinds of parasites” (equine worms, for example). “Here you have a ‘bug’ that is producing a useful molecule grown on a huge scale, and intensively so.” Once the genome was sequenced, he says, “Just looking at it casually, you could see where Ivermectin is made, but you could also see [other gene sequences]—over 30 of these clusters—that it seems should each make a small molecule. We know only three molecules that come from that bug,” he adds—one of them the source of Ivermectin. “That means we know only 10 percent of what it can potentially make.”
But probing those secrets is far from easy. When grown in a lab in pure culture, microbes apparently don’t need to activate all their genetic machinery to survive. In their natural setting, by contrast, microbes live in a complex ecology: they interact with their environment and with other microbes by using a vast array of virtually unknown small-molecule products. These organic compounds often play multiple roles: a small molecule used for signaling among bacteria engaged in mutually beneficial metabolite exchange (one microbe’s metabolic waste is another’s meal) might also be used to kill competitors trying to gain a foothold in the same ecological niche. Such compounds, if researchers could identify them, produce them, and figure out how they work, might form the next generation of medical antibiotics.
At Harvard, an MSI-facilitated collaboration between Kolter and Clardy uses creative methods for prompting even unculturable microbes to yield such genetic secrets. Clardy was among the earliest researchers to use “metagenomics,” which involves sampling the environment—your gut, a lake, or in his case, the soil—and collecting the “metagenome,” or composite genome, of all the individual organisms dwelling there. After extracting the many strands of DNA, Clardy screens for sequences that make compounds. He can isolate these sequences and transfer them into E. coli (or another “tame” bacterium that he knows how to cultivate) in order to get them to express the target compounds. Kolter, meanwhile, has been creating controlled ecological settings, exploring whether certain bacteria behave differently in the context of other species, as they do in their natural environment. He has given Clardy a tool that indicates when a bug responds to a microbial signal, and Clardy has developed other tools for figuring out what that signaling compound is. These organic molecules, often antibiotic, frequently “turn out to be something new that nobody has ever seen before,” says Kolter, “often with properties that are chemically very interesting.”


Photographs by Ann Pearson
Left: The double membranes of Gemmata obscuriglobus may be durable enough to produce “molecular fossils”: traces of early life on Earth that have been preserved for billions of years. Right: Bacillus subtilis forms spores (in green) with lipid membranes that help protect the spore from oxygen damage. These tough cellular membranes appear in the fossil record at about same time Earth’s atmosphere became enriched with oxygen, and may help trace changes in the planet’s early atmosphere.
To show that their method works in principle, the team in 2006 published an early discovery using the bacterium Bacillus subtilis, which has been studied for more than 100 years. The compound they identified, bacillaene, may or may not turn out to be useful in medicine, they caution. But it is a highly complex, resource-intensive structure for any single-celled organism to make, and that suggests that it plays an important biological role. Two new MSI postdoctoral fellows arriving at the medical school this year will use the collaborative techniques Kolter and Clardy have developed to facilitate further discoveries. They will enter a universe of seemingly endless therapeutic possibility, between the small-molecule products of bacteria yet undiscovered, and the 90 percent of untapped coding sequences from the bugs we thought we knew.
Bacterial Biomarkers of Ancient Earth
Within FAS, Ann Pearson, Cabot associate professor of earth and planetary sciences, also became interested in bacteria for what they might reveal about Earth’s early history. Pearson, a biogeochemist, has collaborated with Richard Losick, who during the last 35 years has elucidated in great detail the molecular mechanisms that control life cycle and differentiation in the bacterium Bacillus subtilis. She “does wonderful research,” Losick says, “on [the origins of] biological molecules that can be recovered from hundreds of millions, if not billions, of years ago, reflecting the earliest evidence for life on Earth.”

Roseobacteria, which form pinkish colonies, aid in the formation of clouds by producing gases that nucleate water droplets (detail below).

“If you have very highly preserved, organic-rich, sedimentary rocks that haven’t been deeply buried and heated in their geologic history,” Pearson says, “you can extract measurable amounts of lipids or fats that make up some cell membranes. We call these ‘molecular fossils’ because they have structure, they can be identified, and occasionally you can tell what type of organism they might have come from—for example, there are certain molecules that are produced only by archaea.”
“Ann had the idea,” Losick explains, “that maybe we could learn something from similar molecules found in contemporary microbes.” It turned out that the bacterium that Losick’s lab studies has genes similar to those known to produce a certain type of these molecular fossils. Their shared MSI-sponsored postdoctoral fellow, Tanja Bosak (now an assistant professor at MIT), learned the microbial genetics techniques Losick’s lab uses and obtained evidence that in the contemporary bacterium, these molecules help to protect its spores from damage by oxygen. (Although many life forms need oxygen, it can also cause a lot of damage.) Bacillus subtilis, Losick explains, creates a “particularly macho type of spore, maybe the most sturdy kind of dormant cell on the planet. It can survive extremes of time and temperature and radiation.” What might this molecule have been doing two billion years ago? “The exciting implication,” Losick replies, “is that these molecules may have appeared as a response to the rise of oxygen in the atmosphere, so the timing of their appearance in the rock record may bear on the issue of when oxygen levels first rose.”
“The planet is about 4.5 billion years old,” elaborates another of Pearson’s collaborators, paleontologist Andrew Knoll, Fisher professor of natural history and professor of earth and planetary sciences. “The oldest rocks we can look at are 3.8 billion years old.” Chemical evidence suggests that life was already present then, but certainly “by 3.5 billion years ago there were active microbial communities on the earth’s surface.” These microbes were a little different from those we know today because there was still no oxygen. (Even today, he says, oxygen is just a “veneer on the surface of the planet. If you put your shovel in the mud of a marsh and dig a centimeter beneath the surface you are down to anaerobic microbial ecosystems, which remain critical for most of the biologically important element cycles on the earth’s surface.”) By 2.4 billion years ago, the chemistry of sedimentary rocks suggests that there was “at least a little bit of oxygen in the atmosphere and surface ocean.” Then, “for a period of about eighteen hundred million years, oxygen levels hovered at maybe a couple of percent of today’s levels. Only in the last 600 million years or so do we have environments with enough oxygen to support the biology of large animals, and only in that interval do large animals actually appear,” Knoll says. That is also when the algae first become ecologically important, joining and eventually displacing photosynthetic bacteria as the basis of early food chains in the world’s oceans. “So there is a time coincidence,” he says, “between major events in the history of life and major events in the history of the planet.” Following those threads might also suggest how life could arise, and what it might look like, elsewhere in the universe.

Bacteria fertilize plants through the formation of specialized symbiotic “tissues” on the roots, known as “root nodules.” Bacteria in these nodules form “factories” able to capture atmospheric nitrogen gas and convert it into ammonia-containing compounds that the plants can use for growth. By themselves, plants cannot effect this transformation, which is why they need “fixed” nitrogen (not gas) as fertilizer.
“Everybody is interested in finding the perfect molecule to trace cyanobacteria,” Pearson reports, in order to trace the origins of oxygenic primary production (cyanobacteria use a type of photosynthesis that releases oxygen, and this process is responsible for our oxygen-rich atmosphere). More broadly, she hopes to “figure out how to relate the incredible diversity of microbes that are out there to the kinds of preservable organic molecules—or biomarkers—that they make...and that we can find in the sedimentary record.” Because individual species of microbes are mutable (“They evolve quickly and we have no idea if there is phylogenetic integrity over billions of years”), she is trying to relate particular preserved molecules to their functions in the environment, rather than to species, in order to try to get an idea of what the ecosystem looked like as life evolved on Earth and, in turn, shaped Earth’s environment—oxygenating the atmosphere, detoxifying the oceans, breaking down rock and organic matter to form soils. “MSI has been absolutely essential to building the bridges that I have needed to other research groups,” Pearson says. “Although my questions are geology- and earth-history-driven, the work that I do in order to answer those questions is all biology and chemistry. I don’t practice field geology,” she says, “so building bridges with other parts of the University has been critical.”

Photosynthetic cyanobacteria form colorful mats in the warm springs at Yellowstone National Park, a place that plays a central role in microbial-diversity studies.
Historically, says Knoll, “there have been a few of us in FAS who have been interested in microorganisms, but there wasn’t anything as tangible as a Museum of Comparative Zoology, which has provided a focal point for people interested in animals for 150 years, [or] the Herbarium, which has done the same thing for people interested in plants” (which Knoll calls, half-joking, “green algae with a graduate degree”). He continues, “Because of Harvard’s good fortune in the nineteenth and early twentieth centuries, we built up these really quite remarkable facilities for studying plants and animals...and, not surprisingly, we then populated our faculty with people who study [them]. These areas are not unimportant now,” he explains, “but so many horizons have opened up for studying microorganisms that the simple identification through the MSI of a community of people concerned about microbiology has been tremendously important. Many of the basic biological processes that underlie the nature of changes in the environment are microbial, so in a world where the environment may be changing faster than our ability to understand it, having a better process-oriented idea of what microbial communities are actually doing in nature has practical importance as well.”


Left: These cave mineral deposits were difficult to explain through purely geologic processes. Spelunker scientists in the 1970s discovered that microbes play a role in their formation. Right: The recognition that microbes can respire insoluble metals has led to a complete reassessment of processes, such as rusting, that used to be considered largely abiotic (nonliving) and are now known to have a microbial component. Scientists refer to this as “microbial-assisted corrosion.”
For his part, geochemist Daniel Schrag, professor of earth and planetary sciences and a member of the MSI steering committee, emphasizes that “almost all the reactions on the earth’s surface are catalyzed by microbes—in soils, in waters, in swamps. If you don’t know what microbes are doing, you don’t really know what is going on.”

Photograph by Colleen Cavanaugh
Other kinds of “extremophile” microbes can live without light or oxygen near hydrothermal vent chimneys. The ecosystems around these vents are entirely supported by both the free-living and the symbiotic bacteria.
“It is clear that this field will not reach its maturity, in the sense that it becomes less exciting, during my research lifetime or probably the research lifetimes of my students,” Knoll says. “We have whole new horizons in the nature and treatment of disease; whole new horizons in simply understanding the diversity of life as it actually exists—not what we thought existed because we could see it; whole new horizons in the nature of the relationship between organisms and the environment both today and in the future, and on the timescale of the whole development of the planet. These are wonderfully large, exciting questions and MSI gives us a forum to discuss them. You do find, every once in a while, someone who has actually thought about the same problem in a very different way”—and that can be the most important sort of catalyst: the kind that leads to new discoveries.
All images courtesy of Roberto Kolter, unless otherwise noted
Jonathan Shaw ’89 is managing editor of this magazine.