How did life begin on Earth? On a young, rocky planet, how might chemicals have come together in just the right way to form the very first cells? How did those primitive cells start behaving like life: growing, dividing, and passing on advantageous traits to the next generation?
The origins of life are especially murky because the geological record—the layers of rock and embedded fossils that hold clues about the history of Earth and life—disappears at roughly 3.9 billion years ago, erased by movements in the planet’s crust. As a result, scientists lack direct evidence for conditions on early Earth, including proof of the molecules that might have swirled in primordial ponds and formed the building blocks of life.
This presents a host of questions, precisely the kind of big questions to which Jack Szostak is drawn. He and others believe they can reconstruct in the lab the long pathway that led from chemicals in space, to Earth’s formation, to pre-life chemistry on the planet, to early protocells, and finally to advanced cells with metabolism and protein synthesis. Sprawling explorations like these require expertise in many fields, including chemistry and biochemistry, geology and geophysics, and astronomy.
Szostak (pronounced shah-stak) may be the ideal person to pursue answers. A Nobel laureate, professor of genetics at Harvard Medical School, professor of chemistry and chemical biology in the Faculty of Arts and Sciences, the Rich Distinguished Investigator at Massachusetts General Hospital (MGH), and a Howard Hughes Medical Institute investigator, he is described by others as a brilliant, driven scientist. But he’s also known for mild-mannered humility, including a willingness to dive deeply into subjects that are new to him, and for his collegiality, for helping to foster idea-sharing that is moving science forward.
His own research focuses on one segment of the pathway to life: the protocell, “a really, really simple primordial cell that could assemble from chemicals that were around early on, on the surface of Earth,” Szostak explains. He hopes to understand how it would grow and divide and start to replicate, and eventually evolve. “We may not know what actually happened, but maybe we can work out different possible paths,” he says. “All we can do is try to assemble things in the lab that seem plausible.”
“Directed Evolution”
This August marked Szostak’s fortieth year at Harvard and thirty-fifth at MGH (where his lab is located), a tenure marked by important discoveries in a surprising variety of fields. In the 1980s, his lab conducted experiments with yeast to understand the genetics and biochemistry of DNA recombination—work that led to the double-strand-break repair model, which describes how long strands of DNA break, swap segments, and then rejoin. This prompted subsequent research on the mechanism of recombination during meiosis, the cell division that leads to sperm and eggs.
During the same period, his team also made important discoveries about telomeres—the protective caps, found at the ends of chromosomes, that ensure that DNA replicates properly as cells divide. For this research, Szostak later received the 2009 Nobel Prize for Physiology or Medicine, which he shared with researchers Elizabeth Blackburn, Sc.D. ’06, now emerita at UC, San Francisco, and Carol Greider of Johns Hopkins.
By the time Szostak received the early-morning phone call from Sweden in October 2009, he had already spent more than 20 years making fundamental contributions in other areas of science. After the telomere discoveries in the mid 1980s (research linked shortened telomeres to many diseases of aging), many scientists entered the field and Szostak chose to change direction. “It was pretty clear what the next experiments had to be, and it felt like anything we did would get done anyway,” he recalled. “I’ve never felt that there’s much point in doing stuff that’s going to get done anyway. So that really made me look around and think about what other kinds of scientific questions I could start to address.” In 1984 he accepted an offer to move his lab from what was then the Sidney Farber Cancer Institute to MGH, to join the researchers there working on basic science. “It was an amazing offer: all my research would be fully funded for 10 years,” Szostak recalls. “It was perfect for me since it allowed me to change direction without worrying about writing grants in a new field.”
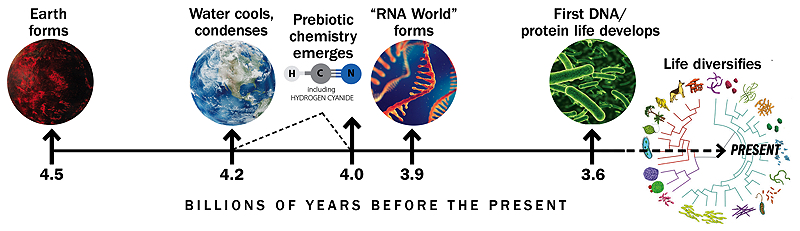
Timeline based on similar graphic by Gerald Joyce
After considering his options, he settled on RNA enzymes known as ribozymes, a field that he saw as “interesting, tractable, and not highly competitive.” He and his graduate students began developing tools to evolve RNA, the single-stranded molecules in cells that copy genetic information contained in DNA. In test tubes his team nudged RNA to take on new roles, such as recognizing target molecules and catalyzing reactions. Known as “directed evolution,” this process involved introducing mutations into the RNA strands, looking for variants that could perform useful functions, and allowing those novel molecules to reproduce. They also did similar work with DNA, peptides, and proteins.
In 1994, Szostak received the National Academy of Sciences Award in Molecular Biology, along with researcher Gerald Joyce, now of the Salk Institute for Biological Studies, for simultaneously but independently developing in vitro evolution of RNA. “It’s a technology for making molecules that do your bidding,” Joyce explained in an interview, describing it as similar to the way agricultural scientists breed cows to produce more milk, or develop crops that resist drought. “This is the molecular version of that. And it’s something that’s now very widely practiced,” he adds (see “Harnessing Evolution,” January-February 2017, page 15), a way of developing new molecules for a range of uses, including medicines.
For Szostak, the work on directed evolution raised new questions. “I got more and more interested in how evolution got started all by itself on the early Earth,” he recalls. “It’s one thing to impose selective pressures and do Darwinian evolution in the lab, where you have enzymes and students and instruments. But somehow Darwinian evolution got started all by itself.” Given that the ability to evolve is a key characteristic of life, Szostak was asking one of the fundamental questions of science: How did life get started?
Origins: “Three Big, Fundamental Questions”
The Canadian-American Szostak was born in London, where his father was studying for a degree in aeronautical engineering. The family eventually returned to Canada, where they lived in Ottawa and then Montreal, and his father worked for the Canadian Air Force. (Szostak’s accent retains hints of his Canadian heritage.) His mother worked for many years in administrative roles for an industrial chemical company, and Szostak held his first summer job there as a teenager, testing color fastness in the company’s dye laboratory. “The job was repetitive and boring, but it did give me my first view into how important it is to test and retest products for real world use,” he remembers. By that time he was “seriously interested” in science, math, and engineering. He earned his doctorate in biochemistry at 25 from Cornell, and calls his adviser, Ray Wu, an important mentor: “He created a great lab environment, but also showed me how to get help on a project when facing problems.”
Although Szostak has conducted some practical, applied research in his career—one of the companies he launched, Ra Pharma, has discovered a drug for the disease myasthenia gravis that is set to begin phase III clinical trials—he is most passionate about basic science.
“This is an exciting time to go into neuroscience because problems can be addressed now that you couldn’t even think about 30 years ago.”
“To my mind there are three big fundamental scientific questions that are super interesting: the origin of life, the origin of the universe, and the origin of the mind or consciousness,” he offers, sitting in his quiet, nearly empty office in the Mallinckrodt Laboratory on Oxford Street in Cambridge. (The office is for occasional meetings; Szostak works mainly in his lab at MGH.) After the origin of life, the origin of the mind interests him most. In the 1980s, when he was planning what to do after his telomere research, he contemplated shifting to the study of neuroscience and even sat in on Harvard seminars on the topic. “It was fascinating, but also depressing, because the technology was so primitive,” he recalls. He has watched with interest as the field’s tools have advanced since then. “This is an exciting time for young people to go into neuroscience because with all the new technology, there are problems that can be addressed now that you couldn’t even think about 30 years ago,” he says. “Yet the overall problem is still so huge and somewhat daunting. The way I look at it, I’m working on the easiest of these big problems.” Because questions about the origins of life are well suited to current research technologies, he adds, he thinks it’s “a solvable problem.”
Model Protocells and “Messy” RNA
Szostak’s team has been making model protocells since the early 2000s, seeking to figure out how they might have assembled and evolved originally. These primitive structures were “extremely simple” in comparison to the simplest single-celled bacterium on Earth today, he explains. Protocells likely included a minimal fatty membrane and initially just one gene that conferred some advantage to the cell. Modern bacteria, in contrast, “have at least hundreds and typically thousands of genes.”
Despite some theories that early life arose near hydrothermal vents in the deep ocean, Szostak is more convinced by research showing that the earliest cells developed on land in ponds or pools, possibly in volcanically active regions. Ultraviolet light and lightning strikes could have helped convert molecules in the atmosphere into cyanide and other useful materials to generate the building blocks of life. The shallow water would give those materials a place to accumulate at high concentrations, and volcanic activity could create hot and cold temperature fluctuations helpful for certain chemical reactions.
A history of Earth and life's beginnings
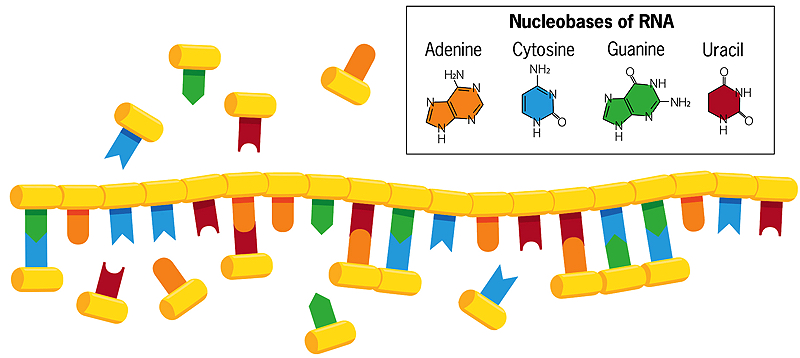
Unlike double-stranded DNA, in which the bases cytosine and thymine pair with guanine and adenine on the opposite strand, the bases of a single helical strand of RNA in water can form associations with free-floating nucleobases (shown in the image above). If these free-floating bases then fuse with each other, a new, mirror copy of the RNA strand is created, which breaks apart from the original when the water is heated. When that new strand replicates in turn, it creates a mirror copy of itself that matches the original strand of RNA. Sometimes errors occur in this copying process, and beneficial mistakes perpetuate themselves. Each chain, acting as a template for its own replication, thus evolves and interacts with its environment.
Some scientists, including Gerald Joyce, suggest that life might have started outside cells, with free-floating molecules encountering each other and forming bonds that would allow them to act like life. But Szostak argues that the cell membrane was necessary, in part because it would keep beneficial genetic molecules together and prevent the useful metabolites made by genetically coded ribozymes from floating away in surrounding water or being snagged by other passing protocells.
Experiments in his lab showed how such a membrane might grow and divide. The researchers combined fatty acids such as oleic acid with water and a buffer (to keep the pH of the solution stable) and then shook the solution. When viewed under a microscope, the ingredients had assembled into vesicles: circular, fluid-filled structures with bi-layered membranes. Adding extra fatty acids to the environment—to serve as nutrients—caused the vesicles to grow long, hair-like filaments so fragile that even a gentle puff of air on the microscope slide caused them to break into pieces. Szostak’s team achieved similar results with different membrane-forming molecules and in different environments, suggesting that this is a plausible way for a protocell membrane to grow and then divide. But how might that structure pass beneficial genetic material to the next generation of daughter cells? “It’s the genetic material that actually looks like a much harder problem,” he says.
To share inherited traits with successive generations, today’s cells rely on DNA—the double-helix molecule composed of the nucleobases adenine, cytosine, guanine, and thymine—to store and transmit genetic information. But replicating DNA in cells requires both the single-stranded molecule RNA and protein enzymes, and genetically encoded proteins are far too complex to have formed spontaneously on early Earth. Because RNA can both store and transmit genetic information (like DNA) and can catalyze chemical reactions (like protein enzymes), many researchers believe that primitive cells used RNA molecules to fulfill both genetic and enzymatic roles.
In the late 1960s, British scientist Leslie Orgel proposed that RNA, or something like it, could have been the first molecule on Earth to replicate and evolve; this became known as the “RNA World” hypothesis. Orgel and others worked for decades to understand how chains of RNA might have come together and replicated, but their efforts were not entirely successful. “There was a lot of progress early on, and then it just stalled because there were a dozen different problems and at the time there was no obvious answer to any of them,” Szostak explains. “Pretty much everybody got frustrated and thought, ‘Maybe life didn’t start with RNA. Maybe there’s something simpler, easier to make, easier to replicate.’”
“We know so much more now, and just by breaking things down into individual, smaller problems, we’ve been able to solve some of them.”
Researchers looked for alternatives to RNA, “and that led to 10 to 20 years of really interesting chemistry, coming up with a lot of interesting molecules,” Szostak notes. “But so far, nothing simpler or better than RNA that really works has come up.” Some researchers support a “metabolism-first” hypothesis, suggesting that life could have started without genetic material, through a series of self-sustaining reactions, but Szostak and others remain unconvinced. About seven years ago, he began revisiting the hurdles that Orgel and his contemporaries faced in understanding RNA synthesis. “We know so much more now,” he explains, “and just by breaking things down into individual, smaller problems, we’ve been able to solve some of them.”
Szostak’s lab now focuses almost entirely on how primordial RNA might have copied itself. Modern RNA comes together in very regular, predictable ways, with nucleotide building blocks clicking together like a chain. Each block contains a sugar (ribose), a phosphate, and one of four nucleobases (also called nitrogenous bases)—adenine, cytosine, guanine, and uracil (usually called A, C, G, and U). The ribose-phosphate units are joined together to make the RNA “backbone.” In modern cells, protein enzymes catalyze the reaction that joins nucleotide units into RNA chains.
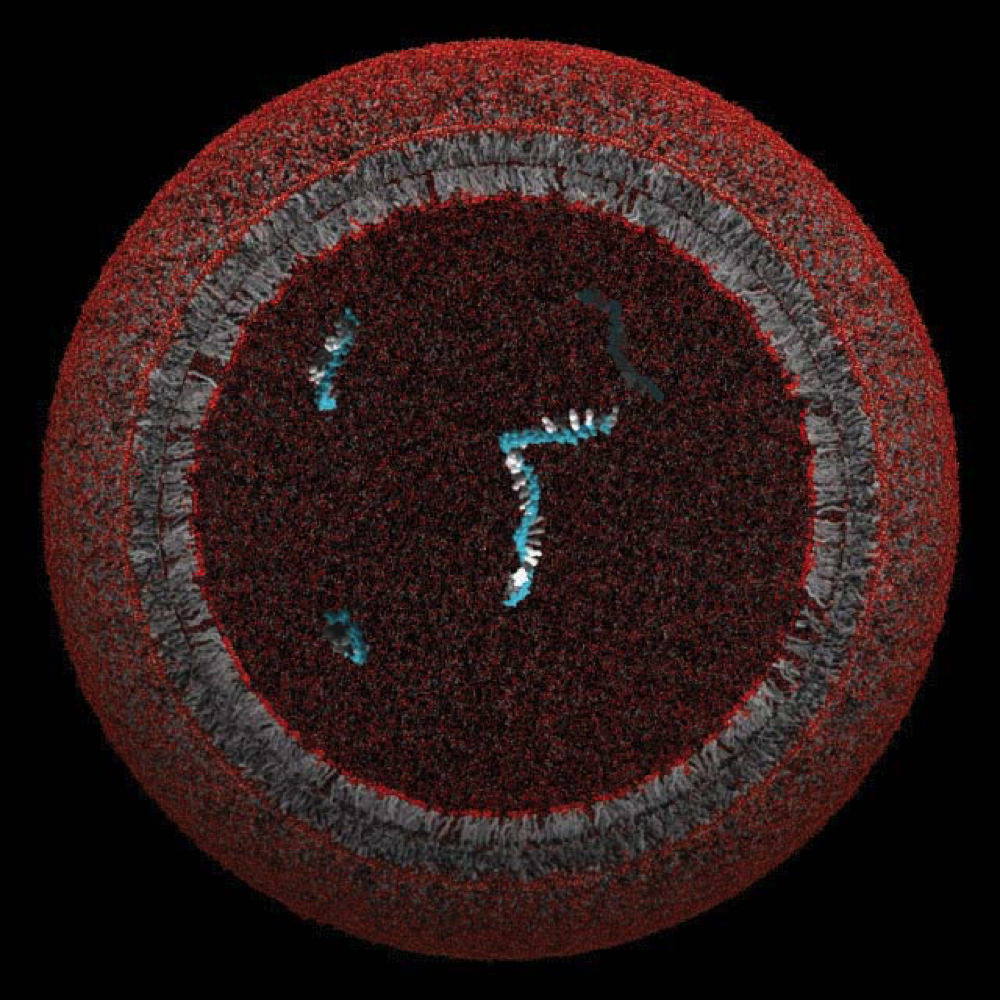
A model protocell, whose lipid membrane might have enclosed and protected a single gene
Image courtesy of the Szostak Laboratory
In protocells on early Earth, the RNA chain would have served as a template on which a new, complementary chain of nucleotides assembled before detaching to become an additional template on which other free-floating nucleotides could click together. But unlike predictable modern RNA, early RNA did not have the benefit of proteins to catalyze the building process. (Because proteins can’t form without the complex and highly evolved cellular machinery required for their synthesis, most researchers believe they were unlikely to exist on primordial Earth.) Early RNA, therefore, was probably messier, with much more variation in the sugar backbone and bases, Szostak says. His team is currently experimenting “to get some idea of what variability would be tolerated and what would be weeded out. Our current model is that you start out with something that’s messy and has a lot of different variations in it, and over cycles of replication, you end up with something that’s closer to modern homogeneous RNA.”
A 2018 paper by Szostak and graduate student Seohyun Kim illustrates the possible variability of early RNA, and its A, C, G, and U building blocks. Scientists have made progress in understanding how C and U could have been generated by prebiotic chemical reactions, but they have struggled with A and G. Szostak and Kim suggest that RNA may have started with different nucleobases, and their experiments have showed that the nucleoside inosine, which can be made from A (adenine), works effectively in place of G (guanosine). “This simplifies the overall problem,” Szostak explains. “Now we just need to know how to make A.”
Other recent experiments in the lab have focused on the metal ions needed to set off the RNA copying process. Researchers typically use magnesium, “but we have to use it at very high concentrations,” which has negative side effects, triggering the degradation of RNA or the destruction of the cell membrane. “Hopefully we’ll find some simple, plausible way of making everything work with less magnesium, or maybe we have to rethink the whole problem and come at it from a different direction,” Szostak explains. “We’re just feeling our way around in the dark, trying to see where there might be a path to a solution.”

Heating and cooling attributable to volcanism, as at Yellowstone's Grand Prismatic Spring, would have facilitated early evolution of RNA.
Photograph by Istock Images
Some of the paths don’t work out, and even yield errors. In 2016, Szostak’s lab published a paper in Nature Chemistry that showed that a peptide could have helped RNA replicate without enzymes. Soon afterward, research fellow Tivoli Olsen joined the lab and could not reproduce those findings. Her review of the paper revealed that the team had misinterpreted the data, and Szostak retracted the paper. “We’re working on hard problems, and the hardest thing in science, as I think Feynman said, is not fooling yourself,” Szostak says. The potential solution was exciting, “and I think it just blinded us to what was going on.” The “saving grace,” he adds, is that they discovered the errors on their own, though he wishes that had happened “before the paper got published instead of after. I’d say a lot of our ideas end up being wrong, but usually we realize that pretty quickly.”
He’s optimistic about the potential of recent discoveries in other labs; for example, John Sutherland of the Medical Research Council (MRC) Laboratory of Molecular Biology in Cambridge, England, recently discovered a new technique for activating nucleotides—chemically modifying these building blocks to power the replication process. Sutherland shared these findings with Szostak’s lab before they were published, and Szostak says they are exploring ways to incorporate this technique into their own experiments.
Once his team assembles working protocells that contain pieces of RNA, they expect information in some specific RNA sequences to confer some benefit to the protocell that surrounds it. For example, previous work suggests that some RNA sequences might fold to become a ribozyme that could make slightly more advanced lipids for the cell membrane. “Any RNA sequence that does anything that helps its own cells to survive or replicate faster will start to take over the population,” Szostak explains. “That’s the beginnings of Darwinian evolution. And then we’re back to being biologists again.”
After winning the Nobel Prize, Szostak could have left the lab to devote himself to travel and speaking invitations, but “He stays focused on the science,” Gerald Joyce says. “That’s what I admire most about him.” Some may see basic research as an intellectual luxury, but its practitioners make the case that all applied science starts with basic science findings. “When Crick and Watson sat down and started making cardboard models of the structure of DNA, they had no idea that it would spawn an industry worth billions of dollars 70 years later,” John Sutherland notes.
Szostak remains committed to chipping away at those big, challenging questions, continuing the work of decades. “I do hope to be able to build an evolving cellular system before I retire,” he says. He’s optimistic about his chances. “I think we’re getting there. There are a few more hard problems, and then I think everything will hopefully be solved in a couple of years.”