Constructing an artificial liver. Altering bacteria to make hydrogen fuel directly from sunlight. Determining how the geometry of damaged heart cells leads to coronary disasters. Creating implantable devices that heal the body by retraining the behavior of cells. All of these projects are the domain of bioengineers, who work at the intersection of the study of life, medical science, and engineering.
Biology and engineering have traditionally represented completely different departments, fields, career paths—even philosophies. But of late, these pursuits have begun to merge in several different ways, making bioengineering one of the most exciting areas of contemporary science. At Harvard, bioengineering offers a chance to bring together the basic studies of life in the Faculty of Arts and Sciences (FAS), the technology-focused work at the School of Engineering and Applied Sciences (SEAS), and the clinically oriented biomedical research of Harvard Medical School (HMS) and its affiliated hospitals. To better position itself as a leader in this growing field, Harvard has been building such connections and recruiting faculty members whose work bridges biology, medicine, engineering, physics, materials science, chemistry, and computer science. Much of the push to expand bioengineering at Harvard comes from students who want to engage in science in a practical way.
In many cases, bioengineers bring not only new tools but a new perspective to traditional biology. “Engineering itself is defined as a field that solves problems,” says Pamela Silver, an HMS professor of systems biology. Traditional biology has emphasized understanding the causes and mechanisms of biological processes, but bioengineers put that knowledge to practical use. Silver says there has been a growing feeling that scientists’ understanding of life, particularly the detailed molecular biology of cells, has progressed far enough that it can become an applied science. At the same time, new technologies and computing power make it possible to transform biology from a “soft” science focused on description to a “hard” science focused on quantifying, predicting, and controlling its properties.
Defining bioengineering can be a challenge because it encompasses a diverse group of research projects that spill across disciplines. In some cases, it involves engineering for biology: the development of new tools to assist biological science and medicine. This area has become particularly important with the rise of small-scale manufacturing and design that make it possible to design tiny devices for sequencing DNA or testing cells for responses to drugs. Another aspect of bioengineering is the engineering of biology. One of the most prominent examples is tissue engineering, which aims to create new tissues and organs outside the body to help patients. But bioengineers are not always working with a medical goal in mind; they also manipulate viruses, bacteria, plants, or animals to act as sensors, waste removal systems, or energy producers. And some bioengineers don’t manipulate living things at all, but use biology as an inspiration for designing new technologies, tools, and products. The following portraits of faculty members suggest some of the University’s growing engagement with bioengineering research.

Photograph by Jim Harrison
Joseph Vacanti and a network of channel designed to carry blood to a tissue-engineered organ, such as a liver.
RECREATING TISSUE
One of the best-known applications of engineering to biology is tissue engineering, in part because pioneering work by Joseph Vacanti and others helped bring its possibilities to public attention. Commercial interest is now rising, boosted by recent advances in manipulating stem cells and regenerating tissue.
During the past two decades, Vacanti’s lab at Massachusetts General Hospital has demonstrated the possibilities of growing tissue outside the body—such as a startling image of a human ear grown on a mouse’s back. Vacanti, who is Homans professor of surgery, got into bioengineering to solve a practical problem that he faced as a young physician specializing in liver transplants: a shortage of available organs from donors. His perspective as a surgeon is aligned with that of an engineer; rather than seeking to unravel all the intricacies of biology, he wanted to find solutions to problems that directly affect patients. But the “simple” goal he pursued—to grow new livers—quickly led to some complex biological problems.
Vacanti credits his direction in tissue engineering to his collaboration, begun 20 years ago, with two other pioneers in biology: the late Judah Folkman, Andrus professor of pediatric surgery and professor of cell biology, a clinician-scientist who launched the study of angiogenesis (the growth of new blood vessels); and Robert Langer, professor of chemical and biomedical engineering at MIT. At the time, scientists could grow cells outside the body, but they amounted to tiny islands of tissue, far short of a functioning organ. The central problem of tissue engineering was clearly one of scale, or, in Vacanti’s words, “How do you make living structures that not only work, but are large enough to help a human?” Multicellular organisms all face a fundamental problem: every single cell needs a supply of nutrients—including oxygen for animal cells and carbon dioxide for plants—and a way of expelling subsequent waste products. “Nature’s solution to that is the design of the vascular circulation,” he says. All multicellular organisms have the same basic vascular design, which involves branching into smaller and smaller scales until each cell is in contact with a vessel that carries nutrients in and out.
“Intrinsic to the solution of scale is the vasculature,” Vacanti says. Building on Folkman’s work, Vacanti and Langer both decided that the key was creating blood vessels in engineered tissue. But growing blood vessels to feed cells takes time, and in the interim cells need an artificial vasculature.
In Vacanti’s office, he holds one example of this principle in the flesh—or rather in the plastic: a whitish polymer dish a few inches square that has had channels as thin as capillaries carved into it by a microfabrication technique similar to those developed for computer chips. The channels are specifically designed to recreate the pressure and flow of a normal vascular bed. Below them is a porous membrane; liver cells can be grown on the other side of the membrane and will then interact with blood flowing through the membrane as they would with normal capillaries. This dish can feed only a sheet of cells, but by stacking multiple dishes together, it’s possible to create something that functions like an organ.
The stack of plastic squares looks very different from the ultimate goal of a liver on demand, but it’s a first step. “Along the way, we decided we might be able to help patients with an implantable assist device,” even if it didn’t look like a real organ, Vacanti says. He and colleagues now hold a patent on their channeled vasculature design. Ultimately, though, he envisions creating scaffolds and vessels out of biodegradable materials that disintegrate in the body after cells populate them, leaving functioning organ tissue behind.
Although initially interested in livers, Vacanti has chosen not to focus on a particular organ, but rather to demonstrate broad principles in several different kinds of tissues. His lab has made achievements in heart, brain, bone, and cartilage tissues as well—demonstrating how tissue-engineering approaches could help diverse patients and different diseases. Though he’s a bit bemused when students refer to his early papers as “classical tissue engineering,” Vacanti’s work has pointed the way toward fresh thinking about how artificial materials can interact with cells.

Courtesy of Dave Savage and Pamela Silver
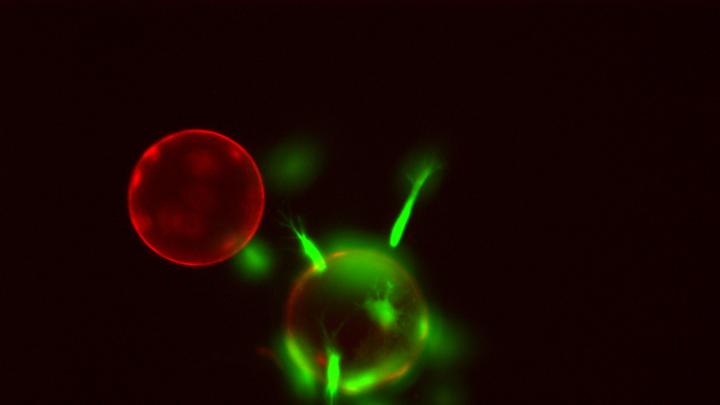
Pamela Silver is working with photosynthetic bacteria—their chlorophyll shown in red and their carbon-fixation machinery in green—with the hope of turning them into “living solar panels.”
REBUILDING LIFE
Pamela Silver has been a leader in the emerging field of synthetic biology, which aims to understand how biological systems are designed in order to apply that knowledge toward redesigning those systems or creating artificial cells and organisms. It offers a new way of using all the detailed knowledge about molecules and cells that biologists have amassed over the past decades: not just to understand life better, but to find ways to put biological components to work for us. Silver says that one of her primary goals is “to make the engineering of biology easier.” Thus her work often focuses on the basic steps needed to manipulate a cell or a system, steps that could be applied for many different purposes.
One of Silver’s major questions is: “Can we engineer cells that will act like computers, that will tell us things?” Her team is working to engineer cells that can relay information about whether they have been exposed to a particular drug or signal, when the exposure occurred, and for how long. As a first step, they created a simple “memory device” and inserted it into yeast cells (and later the cells of mammals), where it reported whether they had sensed a particular signal of interest—sugar in yeast, or a drug in mammalian cells. The device is based on a common phenomenon in biology called a positive feedback loop. The signal of interest activates a protein in the cell, which in turn activates another protein that is capable of turning itself on indefinitely. When the signal disappears, this second protein remains active as a record of the past exposure. Ideally, Silver would like to engineer cells that can provide information about their exposures to DNA damage, drugs, or hormones, and could be used in the laboratory or even implanted in the body as biological sensors. (Her lab also works to engineer individual proteins with the goal of creating better drugs—for instance, by using a knowledge of the biophysics of proteins to modify cancer drugs so that they target tumors without damaging normal cells.)
Synthetic biology can extend from proteins and cells to entire organisms. Plants have long been one of the most attractive subjects for genetic engineering: among the results are organisms that resist droughts, yield larger crops, or serve as sentinels for disturbances or toxins in the environment. One of Silver’s newest interests is using microorganisms to create biofuels that would reduce dependence on fossil fuels. Her lab is now investigating ways to engineer organisms to produce an element that would be useful for fuel, such as hydrogen. They first engineered yeast that can transform biomass into hydrogen, but Silver says, “The real home run would be to get rid of the biomass in the equation and go directly from sunlight to hydrogen.” Her lab is focusing on photosynthetic bacteria, which use sunlight for their own energy. By redesigning bacteria to produce hydrogen or other useful elements from the sunlight, she would like to turn them into “living solar panels.”
Can biology be designed and engineered as easily as computer chips, and can parts of cells or molecules be shuffled around like parts of a machine? Silver’s reference to computers and devices is no accident; synthetic biologists often embrace the language of machines and computer technology. “Some people will say it’s the wrong metaphor,” she says. “I think it’s worth testing.”

Photograph by Jim Harrison
Kit Parker with a cell confined to a triangle (the nucleus and other parts are stained for identification). His lab patterns individual cells onto various shapes to learn how micro-architecture affects a cell’s mechanical properties.
AN ENGINEER’S VIEW OF HEART DISEASE
Kit Parker’s work illustrates how an engineering approach can add a new perspective to biology. Parker, an associate professor of biomedical engineering, focuses much of his research on the structure and function of the heart and how it goes awry in disease. The normal function of the heart relies on several different events: chemical signals within and between cells, electrical pulses of the heartbeat, the mechanical forces of muscle cells as they force blood through the heart.
But rather than viewing disease in terms of genetics or electrophysiology or mechanics, Parker, a physicist by training, sees it as a problem of scale. He explains that events in the heart can happen on different spatial scales, “from protein ensembles on the nanometer level to the whole cardiovascular system, which is on a meter-length scale. That’s nine orders of spatial magnitude.” At the same time, he says, “a protein goes through conformational changes on a nanosecond scale, but people die from ventricular fibrillation in a few seconds. That’s nine orders of temporal magnitude.”
Biology has typically had a much easier time focusing on one scale, rather than drawing connections between scales. But Parker points out that disease doesn’t arise from a single protein or a single cell; instead, it emerges from many small-scale changes. “The question is, what’s the lowest possible level that you can identify disease?” he asks. “And as a biomedical community, are we designing our therapeutics to target that particular spatial scale?” (An inspiration for Parker’s approach was the spectacular failure in the late 1980s of a clinical trial to treat irregular heartbeats with drugs that targeted cell chemistry; the trial was shut down after the drugs led to increased deaths among patients. Underlying the failure of anti-arrhythmia drugs, Parker believes, is an inability to understand how the rapid chemical interactions in heart cells affect the electrical and mechanical forces that control the heartbeat. One of his primary focuses has been to connect the dots between these different events.)
In related research, a team from Parker’s lab, led by Po-Ling Kuo, now an assistant professor of electrical engineering at National Taiwan University, has used a multiscale approach to solve a riddle about the shape of heart muscle cells (cardiomyocytes). Parker explains that pathologists who first compared hearts from people with and without heart disease found that normal heart cells look roughly like oblong cylinders. In hearts that had become thickened because of maladaptive growth, the cells were a shorter, fatter cylinder; those from enlarged hearts had cells that were longer and skinnier. Parker’s group found that “there’s this sweet spot, an optimal shape to get the maximum contraction from a cardiomyocyte.”
Parker encourages his team to apply tools and knowledge from different fields to a problem, rather than forming questions around the technologies at hand. In this case, the researchers used software originally developed to analyze fingerprints to study what was happening at the molecular level in cells. They found that when the overall shape of the cell changes, tiny protein motors responsible for muscle contraction get misaligned. “It’s a stunning thing,” Parker says. “It shows that on the whole organism level, you’re going to die of heart failure because you’ve got a nanometer or micrometer architectural problem inside the cells of your heart.”

Prakriti Tayalia, David Mooney, and Eric Mazur
Cells (green) within a three-dimensional structured polymer. Prakriti Tayalia, mentored by professors David Mooney and Eric Mazur, uses confocal microscopy to show cell infiltration and adhesion in this engineered scaffold.
MASTERING CELLS
Biologists tend to focus on understanding biological processes, but this understanding doesn’t necessarily translate into ways to control disease or repair physical defects. McKay professor of bioengineering David Mooney sees his field as a critical link between traditional biology and new medical advances. Consider the wealth of research during the past two decades that has shown that tumors depend on angiogenesis for their survival, and that blood vessels are critical for growing artificial tissue to repair injured and diseased organs. These discoveries have led to interest in shrinking blood vessels in cancers and promoting their growth in transplanted and engineered tissues. What is often missing, Mooney says, is a systematic study of what chemical or physical factors are needed, in what amounts, and when, to influence the process in a predictable way. “In biology, there’s a real emphasis on discovery,” he says, “not an emphasis on controlling what’s happening.”
So Mooney is working to “design materials that can communicate with the cells in the body and control them.” Unlike Vacanti, whose goal is to engineer therapies that can be introduced into patients as soon as possible, Mooney focuses on pursuing new ideas in tissue engineering that may take many years to realize. An example is the use of stem cells to replenish tissues. Scientists have discovered chemical factors that control how stem cells mature into specific cell types, but they have struggled to use that knowledge to manipulate stem cells predictably. Mooney’s lab works to determine how much of a particular factor is needed, in what location, and when.
The goal is to use this information to create materials and devices that mimic the natural environment of the cell. “Cells actively probe, push, and pull on their environment to understand their surroundings,” he says. Tissue engineering often involves growing cells outside the body on artificial scaffolds that imitate the physical properties of tissues. Mooney would like to make “smart” scaffolds that can communicate chemically as well as physically with cells and release their chemical signals in a controlled fashion over time.
Ideally, he says, cell- and tissue-based therapies shouldn’t necessarily have to rely on implanting cells grown outside the body: it may be possible to control the cells already there. Mooney believes that scientists could create an environment inside the body that promotes cell regeneration or encourages cells to reverse disease. For instance, many cancer scientists are looking for ways to develop a cancer vaccine that could stimulate the immune system to destroy a tumor. Mooney imagines that one could implant a device that can “teach” existing cells to act against tumors—first by attracting immune cells and then secreting signals that cause them to attack cancer cells in the body.
Although biology may have the reputation of being “messy” and difficult to control, Mooney says that it also offers advantages for engineers. “Another way to think of it is that biology tends to be quite robust,” he says. “It has the ability to get to certain endpoints even if there are bumps and noise along the way. The key is to work with that, not against it.”

Joanna Aizenberg
Like tiny flowers, micro-florets created in Joanna Aizenberg’s lab open and close in response to changes in environmental moisture. These structures, their action controlled by a hydrogel “muscle,” can be used to catch and release tiny particles.
MIMICKING LIFE’S DESIGNS
Although many bioengineers use tools of engineering to manipulate living things, Joanna Aizenberg, McKay professor of materials science, works in the reverse direction: she uses biology as an inspiration for engineering new artificial materials. Aizenberg (who is also Wallach professor at the Radcliffe Institute and professor of chemistry and chemical biology; see “Portrait,” July-August 2008, page 59) came to Harvard a little more than a year ago from Bell Labs, where she had developed materials and devices inspired by creatures such as brittle stars (a close relative of starfish), which are covered in a sophisticated array of tiny lenses.
This offshoot of bioengineering—often called biomimetics or bio-inspired design—sees biology as a source of creative ideas. “Through evolution, nature created very sophisticated solutions to complex problems,” Aizenberg explains. These solutions can be found simply by observing the structures washed up on a beach, something she does regularly. As a materials scientist and a chemist, she finds much to admire.
As an example, Aizenberg holds up a long, slender tube: the skeleton of a sea sponge. Though made of a seemingly delicate white lattice of glass, the skeleton is surprisingly strong and rigid. Aizenberg wants to build something with similar properties; she next holds up two cylinders made of orange plastic. One has a lattice of open squares, while the other has a more complicated crossed lattice similar to the sponge’s. The more intricate one is far more rigid than the simpler design. Part of her work is to identify which properties give natural structures an advantage—whether it is the materials or their structure—and incorporate those properties into artificial materials and structures.
Though many of her projects have focused on mineralized tissues like the sponge skeleton and the brittle star lenses, Aizenberg’s newest interest is cilia: hair-like structures that have a variety of roles in many organisms. Hair cells on the outside of an organism can serve as sensors; sea animals can use them to navigate in deep waters even if they can’t see. Within the body, they can help to channel the direction of fluid flow—for example, in blood vessels. The proper movement of fluid by cilia even ensures that our bodies develop the proper left-right symmetry in the womb. Aizenberg believes that cilia-like structures could help solve several engineering problems—for instance, improving the flow of fluid through narrow channels in small devices. Microfluidics—in which fluids are manipulated within areas of a millimeter or less—is a growing field in biotechnology, but moving fluids through increasingly small spaces is challenging. “If we can make channels or pipes with these hairy walls, similar to ciliated walls,” she says, “we can decrease the pressure and therefore the energy needed to push liquids in microchannels, or reduce drag in pipelines.” (Although one might assume that the presence of hair-like projections in a channel would obstruct flow, in fact they repel water so well they actually help speed flow along.)
Aizenberg’s team is currently developing a “nanofur” with hair-like projections that change properties in response to humidity: attracting water when dry and repelling it when wet. The ability to change in response to the environment is one of the properties that make biological materials more useful than artificial ones.
Aizenberg believes that students are attracted to biomimetics because it lets them study nature in a way that benefits real-world problems. It also draws together people from different disciplines, blending knowledge of biology with physics, chemistry, materials science, mathematics—Aizenberg has even worked with archaeologists. “Almost anyone can contribute to it in one way or another,” she says.

Photograph by Jim Harrison
Radhika Nagpal with a sheet of dividing epithelial cells within a fruit fly’s wing. Her lab studies how cell networks form and then influence large-scale properties of tissues.
PROGRAMMING LIFELIKE BEHAVIOR
Assistant professor of computer science Radhika Nagpal also finds inspiration in biology. In particular, she is interested in the complex harmony of biological systems, such as the cells of the heart beating in synchrony or ants cooperating as a unit to achieve a collective task. “You cannot destroy a colony by stepping on a few ants,” she says. “Can we build systems that have that kind of robustness?” Many biological processes rely on coordinated activities among independent individuals acting without a distinct hierarchy or central command; Nagpal believes that computer science has much to learn from this bottom-up approach.
She works with biologists such as Donald Ingber, Folkman professor of vascular biology and of bioengineering, to understand living processes and then looks for ways to apply those guiding principles to the design of computer systems and programmable structures that have properties of living organisms—such as sensing their environment and adapting to it or repairing themselves.
Robotics is an ideal area for creating artificial systems that mimic biological behaviors. Nagpal programs modular robots to work together to imitate the organization of groups of cells and organisms in biology. She says that many different coordinated processes in nature rely on similar principles; the individuals must be able to come to an agreement based on what those around them are doing. “They’re trying to create homeostasis,” she says.
She and her lab members have used these principles to program modular robots to solve problems (see the "Bioengineering in Motion" Web Extra). In one example, a student constructed a table that keeps itself level, even when it is placed on uneven surfaces, by means of cooperation among its separate components. In another, a line of small robots, linked together, are able to grasp a balloon gently by coordinating their movements and pressure. The same principles could be used to build structures that actively respond to their environment, such as a bridge that keeps itself level or a structural support that becomes stronger in response to greater pressure. Nagpal has used similar algorithms to help networked computers keep the same time, and thinks that such techniques could also help computers schedule tasks and relay information over networks more efficiently.
On a smaller scale, Nagpal imagines stents that can adjust their pressure against the wall of an artery. The design of small-scale programmable materials and devices is becoming more feasible as engineers combine tiny sensors and actuators—components that perform actions like shrinking, rotating, or hinging. Tiny technologies like these underlie small devices that sense their environment and respond with appropriate behaviors, such as filaments that shrink in unison, just as muscle cells contract.
Like many other people working in bioengineering at Harvard, Nagpal stresses the importance of finding collaborators and seeking input from colleagues across disciplines. For many bioengineers, in fact, collaboration is a necessity, because specific expertise in biology, physics, or math can be absolutely essential to a project. Her students may present their work at hospitals or math seminars to solicit new perspectives. Within SEAS, she says, “It’s very easy to talk to people and find out what they’re up to, and the connections happen really fast.” At the same time, electronic communication makes it easier to maintain links to other researchers whom she doesn’t see face-to-face.
“I really like all these opportunities, and there’s a lot of informal contribution,” she notes—but the bigger problems are, how many people can one possibly interact with, and what’s the ideal size of a network? In asking these questions, it’s clear that even the living system of scientists at Harvard could serve as an inspiration for Nagpal’s work. This freedom to look for ideas and resources beyond a single discipline—whether the goal is to treat a disease, design a computer network, or understand the structure of cells—is what makes bioengineers across the University excited about the prospects of their field.