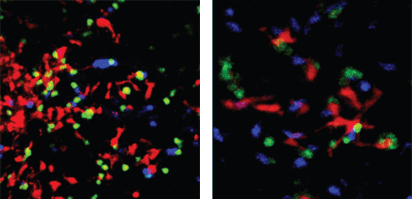
The scenes are familiar from biology textbooks. A long string of DNA is copied to form a matching strand. A virus infects a cell by stealing through its membrane. Two white blood cells meet and confer before launching an immune attack.
In textbooks, all these processes that are so fundamental to the lives of cells are typically depicted in drawings or static snapshots captured by powerful electron microscopes. But that’s changing. A growing revolution in imaging is making it possible for biologists to watch small-scale events as they unfold in living cells and tissues.
“The human brain is vision-focused,” says professor of molecular and cellular biology Jeff Lichtman. “If we see things, then we think we know what they mean.” To be able finally to see events that were known only in theory is incredibly satisfying for scientists. Even more important, this revolution also opens up the possibility of learning things about life that could never be studied before.
Ironically, the technology enabling much of this change is the same one that launched the study of modern biology centuries ago: the light or optical microscope. A congruence of factors has shuttled these instruments back into the forefront of biology in recent years, after almost a half-century during which they were overshadowed by more powerful techniques such as electron microscopy and x-ray crystallography, which are able to create images on the level of single molecules.
New technologies—more sophisticated imaging techniques, fluorescent molecules that act as beacons of light in the cell, and the computing power to gather and stitch together multiple images and create videos from high-powered microscopes—make it possible to harness one of light’s key advantages: gentleness. Unlike higher-resolution techniques, light microscopes can image biological structures without killing them or chemically fixing them. At Harvard, the resurgence of light microscopy is making it possible to see structures and events that have never before been seen in the context of living cells and organisms. New discoveries are emerging at many scales of life, from the activation of a single gene in DNA to the development of disease in an organ.
Pushing Microscopes Further
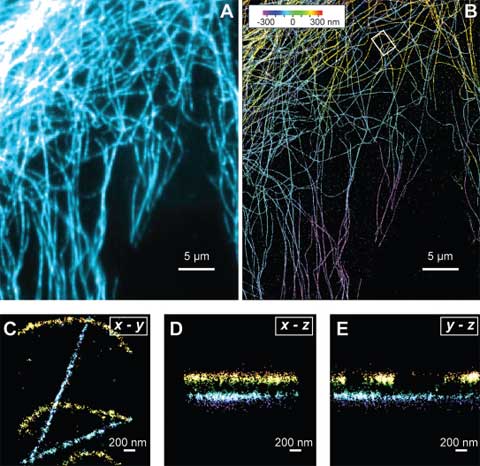
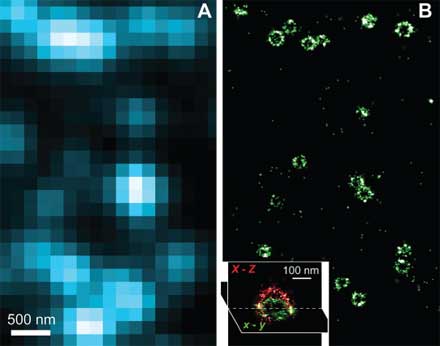
At the molecular scale, Xiaowei Zhuang, professor of chemistry and chemical biology and of physics, is pushing the boundaries of what light microscopes can capture. A typical light microscope can easily image a single cell and some internal structures, but most other objects—viruses, clusters of proteins, DNA—cannot be seen in great detail. That’s because these smaller details lie within light’s diffraction limit—the point at which light waves begin to interfere with one another, blurring the image. The question of how individual molecules in cells interact is fundamental in biology, but for the most part these interactions lie beyond the reach of light microscopes. Zhuang’s lab has been working on a new “super-resolution” optical imaging method that uses clever tricks to image objects a tenth the size possible using normal light microscopy.
Like many new optical imaging techniques, Zhuang’s takes advantage of small fluorescent molecules called fluorophores to create an image. Scientists can determine the location of a single fluorophore under a microscope with great precision, even though it is much tinier than a microscope’s resolution. Light emitted from the molecule will produce a blurry image, but the center of this blur indicates where the actual molecule resides.
The problem, Zhuang says, is that in most biological contexts a scientist would not be imaging a lone fluorophore but many at once, and if these tiny lights are too close together, resolving them is impossible. Her solution is a technique called STORM (stochastic reconstruction optical microscopy), first reported in 2006, which involves switching on only a small number of fluorophores at a time, then iterating the process until the position of each spot has been separately determined. Last August, her team was able to use the technique to produce multi-colored images, making it possible to view several different types of molecules in the cell and watch how they interact. More recently, her team was able to use STORM to produce three-dimensional images—essential for knowing the exact location of a molecule. “We don’t live in a two-dimensional world,” she says. “If we want to see how molecules interact unambiguously, we have to have the third dimension.”
So far, Zhuang’s team has imaged static structures such as a piece of DNA or part of the cell’s inner skeleton; she says it will take some work to make STORM broadly applicable for imaging more dynamic events. “The method doesn’t just give you high resolution for free—what is sacrificed is speed,” she explains. STORM uses the dimension of time to isolate different parts of an image and then stitch them together. Currently the technique takes tens of seconds to capture a high-resolution image, whereas many cellular processes occur within seconds or fractions of seconds. But Zhuang’s technique, despite its limitations, has generated excitement, and she has been overwhelmed with requests for collaborations. For instance, in collaboration with Catherine Dulac’s lab (in the department of molecular and cellular biology), her team is beginning a project to look at chemical communication that takes place in the synapses between nerve cells. “The synapse itself is just about the size of a diffraction-limited spot,” Zhuang says. A closer look “really calls for higher resolution.”
One Molecule at a Time
Zhuang tackles the resolution problem by switching on fluorophores one at a time, but professor of chemistry and chemical biology X. Sunney Xie focuses on molecules that are naturally isolated from others of their kind. He says that many questions in biology can be answered only on the level of single molecules. An example is the process of gene expression—how the DNA template of a gene is turned into messenger RNA and then into proteins. A given cell may contain only one or two copies of a given gene, and only a few copies of the messenger RNAs created from each gene. Because imaging is the best way to study singular events, it serves as the cornerstone of a growing field of “single-molecule” studies, which investigate how individual molecules and groups of molecules behave and interact.
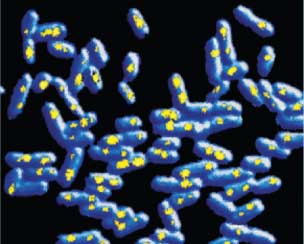
Images courtesy of Gene-Wei Li and Peter Sims, Harvard University (top) and Yuichi Taniguchi, Harvard University (bottom)
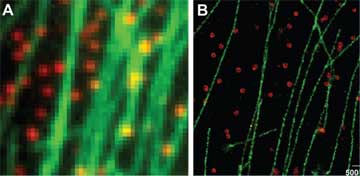
The unique features of DNA molecules are ideal for a strategy that Xie developed called “detection by localization.” If a fluorescent molecule appears only in one place, it can easily be detected over the strong background of fluorescence that cells naturally emit, especially if the molecule is attached to a relatively stationary structure such as DNA or the cell’s outer membrane. Because different pieces of DNA have unique sequences, Xie’s group creates fluorescent molecules that appear only at a single spot on the genome. Isolated, these fluorophores can be located more precisely.
Previous single-molecule experiments relied on molecules taken out of cells, but Xie’s group has been imaging events related to gene expression in live bacterial cells. For instance, his group has been able to visualize the behavior of a type of protein called a repressor, which regulates gene expression. The transcription factor itself is kept inactive by a protein called a repressor. By labeling a repressor with fluorescence, the team could observe it binding to a unique target among vast stretches of DNA. But when they added a chemical signal that prevents such proteins from binding to DNA, they quickly dispersed into the cell. When the signal was removed, the repressor found its target again in about a minute through a process of trial and error. After proving they could use these tools to study singular events in the cell, Xie’s group is applying the technique to investigate other steps in gene expression, as well as DNA replication and the repair of mutations in the genetic code.
Molecules In Motion
High-resolution microscopes are enabling scientists who have long studied cellular processes to put static pictures of cellular structures into motion. Tomas Kirchhausen, professor of cell biology at Harvard Medical School (HMS), has for decades studied how things get in and out of cells, from viruses and bacteria to small molecules and hormones. This cargo, whether intruders or simply chemical signals from surrounding cells, is captured and taken into the cell by capsules called vesicles.
Kirchhausen’s lab has performed a great deal of work to understand the precise structure and function of clathrin, a protein that helps deform the cell membrane as it forms the outer coat of vesicles. Other imaging methods yielded detailed pictures of clathrin and the lattice-like coats it forms, but these were only snapshots, he says, whereas “light microscopy allows you to figure out how things are happening.” In the past few years, his team has begun to capture movies of vesicles as they form at the cell membrane and capture incoming cargo, then break off from the membrane and dance into the cell interior.
When they first began collecting images, Kirchhausen and his colleagues could capture only one frame every few seconds; now they can collect images in milliseconds, creating real-time movies. Although vesicles are smaller than the resolution of the microscopes he uses, Kirchhausen says it’s enough to be able to detect the presence of each one when it is lit up with fluorescent molecules: “We cannot see the actual shape of the object, but we can see its properties—where it is, where it goes, when it forms, and when it disappears.” By pairing the dynamic nature of light microscopy with the detail of higher-resolution methods, his team can now answer questions that confounded them in the past, such as how different chemicals or genetic alterations affect vesicle traffic. The movement of cellular cargo is fundamental for a cell’s survival and communication, but it also underlies viral infections, cancer, and neurological diseases.
Cells In Context
Zooming out to the scale of cells, new imaging techniques are helping scientists see how these building blocks behave in their natural context—within tissues in the living body. This is particularly important in the brain, where billions of neurons gather together in an intricately connected web. It is impossible to know how the brain works by studying neurons in a culture dish; their relationship to neighboring cells in the brain dictates their function.
Jeff Lichtman’s laboratory has developed techniques for observing the web of connections in the living brain and watching how they change over time. Lichtman studies how connections among neurons form in early life. In humans and other mammals, neurons send out many branches to other cells early in the nervous system’s development, but then, Lichtman says, “a lot of the connections that were made are pruned away.” He likens the process to a massive competition for territory and connections among individual nerve cells. “When you have this pruning going on,” he says, “many branches from different nerve cells connect to the same target, and then they compete with each other.”
If every nerve cell emits the same color of fluorescence, he notes, “you’re kind of lost. You can’t trace them all back.” In partnership with the lab of professor of molecular and cellular biology Joshua Sanes, Lichtman’s team was able to engineer mice with neurons that glow with an array of different fluorescent colors. A genetic technique randomly shuffles different combinations of fluorescent proteins in red, green, and blue to give each cell a unique hue. The approach is akin to color-coding electrical wires, so each one is identifiable in a tangle. With these “Brainbow” mice, Lichtman and his team can trace the fate of a single nerve cell over time and across distances. Lichtman’s goal is to use these mice along with higher-resolution electron microscopy to trace the development of entire networks of neurons, beginning with the relatively simple peripheral nerves that project to muscles in the neck.
How do networks of interconnected neurons work once they’re in place? In the past, neuroscientists studied the activity of neurons in the brain by measuring their electrical activity. R. Clay Reid, professor of neurobiology at HMS, who studies the brain’s visual system, said that until five years ago, imaging was not even a major part of his work. “We used to put tiny wires into living brains to eavesdrop on one or two or maybe 10 living cells,” he says. When cells were active, they “fired” an electrical signal that could be measured.
It was through this sort of painstaking measurement that neuroscientists mapped out the function of different areas of the brain’s cortex. Reid explains that in the 1980s, for instance, Margaret Livingstone, a professor of neurobiology at HMS, worked with Nobel laureate David Hubel, now Enders professor of neurobiology emeritus, to discover that certain regions in the visual cortex are important for color vision. But during the past few years, light microscopy techniques have made it possible to monitor the activity of the individual brain cells within an entire region of the brain simultaneously. “Now we can actually see all of the cells,” Reid says. “It’s really exciting.”
The key is calcium: when a neuron “fires,” calcium molecules rush into the cell. Using a fluorescent reporter that lights up with this calcium influx, scientists have a visible proxy for the brain’s electrical activity. Reid and the members of his laboratory can now watch the activity of thousands of cells at once, and they can also determine a single cell’s function with astounding precision. Two neighboring neurons may have branches that are tangled together, and yet respond to completely different visual signals. Reid’s team can isolate which cells, for instance, respond when an animal views lines oriented in one direction versus another. He believes it will be possible someday to have a detailed map of the entire circuitry of the visual system, and his group is teaming with Lichtman’s lab to explore ways to chart networks in large regions of the brain.
Cells on the Move
Light microscopes are particularly useful in capturing dynamic events, and perhaps no cells are as dynamic as blood cells. Ulrich von Andrian has been tracking their behavior in the past several years, focusing in particular on the 500 billion T cells of the immune system. Von Andrian, the Mallinckrodt professor of immunopathology at HMS, specializes in “intravital” microscopy, which images events in living animals. The techniques help track how these “immune cells” migrate throughout the blood and lymph vessels of mice, and how they collect at peripheral sites in the body—whether beneficially, in response to an injury or infection, or pathologically, as in the case of autoimmune diseases. To capture very fast events such as the movements of these blood cells, team members record videos under a stroboscope, just as photographers use strobe lights to capture the trajectory of a bullet. For slower events, they use high-resolution two-photon microscopes that capture several frames per minute, which are then speeded up to make a time-lapse video.
These techniques let von Andrian’s team understand immune-cell behavior better: for instance, how T cells are activated in response to a foreign particle or antigen. The vast majority of T cells circulating throughout the body are inactive, or “naïve”; they rouse into action only when they meet a very specific foreign antigen presented on the surface of another immune cell. As von Andrian explains, this process was thought to be fairly static—when the T cell met the right antigen, it would bind to the antigen-presenting cell for a number of hours, and eventually become active. In fact, his group found that T cells rove from one cell to another in a fashion he compares to bees moving from flower to flower, rather than staying on the first cell they find.
A recent study offers a possible explanation. T cells, he and his colleagues found, actually seem to sense how much of a particular antigen is present. The cell is “collecting information, and it somehow remembers what it has seen in the previous hours,” von Andrian explains. Activation requires “a cumulative signal from many different encounters,” and not just a single match.
Such nuances would be difficult to determine without seeing them directly. But von Andrian emphasizes that research does not end when a video is captured. “One danger of imaging is that you just sort of stand in awe in front of the pretty pictures and the beauty of it all,” he says. “It’s easy to forget about the mechanism and the ‘why’ of it all.” Von Andrian pairs imaging techniques with the classic approach of a biologist: to come up with a hypothesis and test it, often through genetic or chemical manipulations. Imaging lets scientists see how any change—a genetic defect, a drug, an infection, or a wound—affects the immune system at the level of its individual cells.
Imaging the Body
Fluorescence microscopy offers the opportunity not only to track specific molecules in cells, but to see how those cells behave in an entire organ or part of the body. Charles Lin, a member of the Center for Systems Biology (CSB) and the Wellman Center for Photomedicine at Massachusetts General Hospital (MGH), develops novel methods and imaging equipment to follow cells that move throughout the body. “The holy grail is if you could follow a single cell through the entire body, and its interaction with the environment,” he says.
His work, which he says was inspired by von Andrian’s intravital techniques, includes studying aspects of the immune system, cancer, and stem cells, and how they intersect. Why, for instance, do tumor cells begin to metastasize and migrate throughout the body? “If you figure out how cancer cells spread, and interfere with the signals of spreading, then you can reduce metastasis,” Lin explains. Unlike most labs at the medical school, which rely on ready-made equipment to pursue research, his group focuses on developing new imaging devices for specific applications—to access a certain part of the body, for instance, or to find very rare cells such as blood stem cells after a bone marrow transplant.
Lin says that, as a technology-focused group within a hospital, the center can help adapt new microscopic techniques, increasingly making microscopes—those hallmarks of the laboratory—more useful for the clinic. There, most imaging methods, such as MRI, CT, PET, and ultrasound, detect physical properties of tissues in order to create images (see glossary below). But to track and study specific molecules, light microscopy has the advantage, says Ralph Weissleder, professor of systems biology and radiology at HMS. Weissleder oversees the CSB and the Center for Molecular Imaging Research at MGH. “What we hope to do at the end of the day,” he says, “is to understand biology as it unfolds in vivo rather than in snapshots.”
His group works on developing new probes that have the ability to fit, like a key into a lock, into a pocket on the surface of a protein. A fluorescent molecule attached to the probe acts as a “reporter” that lights up proteins for the microscope. Weissleder says that the newest probes are constructed to fluoresce only when they have reached their targets, which results in a more distinct signal.
Using this technique, scientists can study and monitor the course of diseases in living animals. With cardiovascular disease, for instance, arteries become inflamed, which can eventually lead to a blood clot and a stroke. A probe specific for inflammatory enzymes can light up areas where the risk of clotting is greatest. These imaging tools could help scientists and clinicians monitor a disease and track whether a medication or other treatment is working.
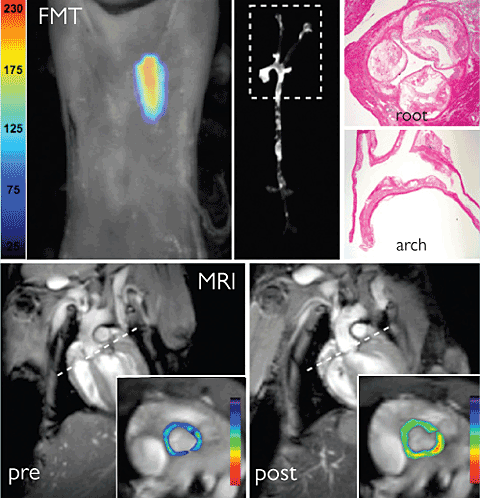
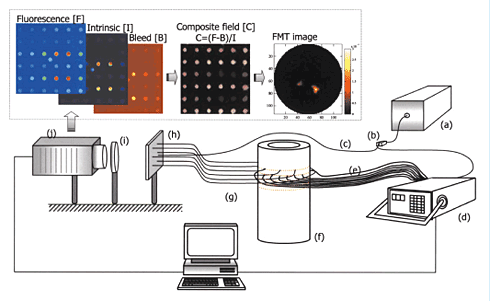
Most microscopic techniques are limited by depth; looking deeper into the body usually requires a surgical incision to gain access because it’s impossible, at this point, to image fluorescence in the entire body at once. But a recent technology developed at MGH makes it possible to do just that—at least in small animals like mice. The technique, called fluorescence molecular tomography (FMT), creates a three-dimensional image of fluorescent proteins inside a living animal. The technique uses light itself to gather information about the tissue and reconstruct an image, similar to the way hospital CT scanners use x-rays. An animal with a fluorescently marked object inside it, such as a tumor, is exposed to beams of focused light at varying angles. At the other side, a sensitive camera records both the transmitted light and fluorescence from inside the body. A computer can use information about how the light beams are scattered by tissue to calculate the depth and degree of the fluorescent signals.
While there may never be a “whole-body” microscope that can scan a patient’s insides the way an MRI can, Weissleder believes that techniques like FMT and fiber-optic microscopy will prove useful for specific applications. Thus FMT is being developed for clinical use to visualize inflammation in the carotid artery in patients with atherosclerosis. Weissleder says that the key barrier to bringing FMT and other microscopy techniques to the clinic is getting the probes approved for use in humans, but he notes that probes are beginning to enter clinical trials. His group is also investigating nanoparticles that can be detected by different types of imaging techniques, such as MRI and PET, as well as optical microscopy. He believes that optical methods won’t replace these standard techniques, which still offer the best way to see inside the entire body, but says they can add molecular information to that obtained from existing technologies.
Seeing Single Cells, Counting Masses
The resurgence in imaging excites biologists for two reasons: it allows them to see individuals, and it allows them to count the masses. Being able to watch and track a single molecule, cell, or process offers a much more complete picture of how life works. Traditionally, many processes were studied using large numbers of molecules or cells to get an average result. But Sunney Xie points out that in events like gene expression, “you cannot synchronize the activities of each cell. It’s stochastic.” Stochastic events involve an element of chance—no two cells will be exactly the same. Xie points to a photograph of his twin daughters to illustrate the idea; they have identical genes, but distinct personalities.
Being able to view singular events also gives scientists a new point of view. In the Brainbow mice, for instance, it’s easier to visualize how each cell competes with others. “Each cell is an individual and each is trying to make a living,” Jeff Lichtman says.
While the view of the singular is thrilling, scientists are equally excited by the ability to quantify many objects and events at once. Cells and molecules labeled with fluorescence can be counted, which means that scientists can see whether their numbers wax and wane as events unfold. Charles Lin explains that when studying whether a particular treatment keeps tumors from metastasizing, it’s important to be able to count the number of cells traveling to other areas of the body to know if a therapy is working. The ability to put numbers on results is a long-sought goal of biology, which has often been seen as a descriptive science.
Tom Kirchhausen predicts that in the next few years, scientists will use imaging to better understand complex processes such as cell division and the paths that viruses take to cause infection. Like many of the biologists involved in imaging, he is enraptured with what he’s been able to see in recent years. “Someone in the lab had a good result yesterday, and I had trouble sleeping,” he admits. “It’s so exciting.”
Glossary
CARS microscopy: a technique for visualizing specific molecules in cells based on their unique patterns of vibration when exposed to beams of laser light at specific frequencies.
Computed (axial) tomography (CT, or CAT, scan): an imaging technique that uses x-rays at many angles to generate cross-sections of the body, which a computer then stitches together into a three-dimensional image.
Diffraction limit: a limit to a microscope’s resolution imposed by the wavelength of the light used to illuminate a specimen. At distances smaller than about half the light’s wavelength, the light waves interfere with one another.
Electron microscopy: a technique that uses electrons rather than visible light to produce an image at up to a thousand times the magnification of light microscopes.
Fluorophore: a molecule that is able to fluoresce, i.e., to become excited by a particular wavelength of light and then emit light at a longer wavelength as it returns to its normal state.
Fluorescence molecular tomography (FMT): an emerging technique that uses beams of light similar to x-rays in CAT scans to create a three-dimensional image of fluorescent molecules in the body.
Green Fluorescent Protein (GFP): a protein originally found in jellyfish that fluoresces green when exposed to blue light.
Magnetic resonance imaging (MRI): a diagnostic technique that uses a powerful magnetic field and radio waves to generate detailed cross-sectional images of tissues in the body.
Positron emission tomography (PET): a technique for detecting functional activity in tissues by introducing a small dose of radioactive chemical into the body and using a computer to transform the resulting gamma radiation signals into three-dimensional images.
Optical or light microscopy: the use of visible light transmitted through or reflected from a sample and then passed through lenses to provide a magnified view.
Resolution: the minimum distance between two points that a microscope can distinguish.
Ultrasound: a diagnostic tool that uses reflected high-frequency sound waves to examine internal body structures.
X-ray crystallography: a method to determine the structure of molecules by aiming a beam of x-rays at a crystallized molecule and measuring how the rays scatter.