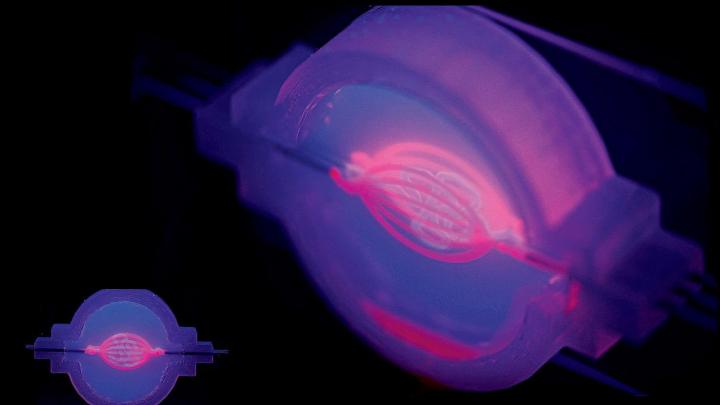
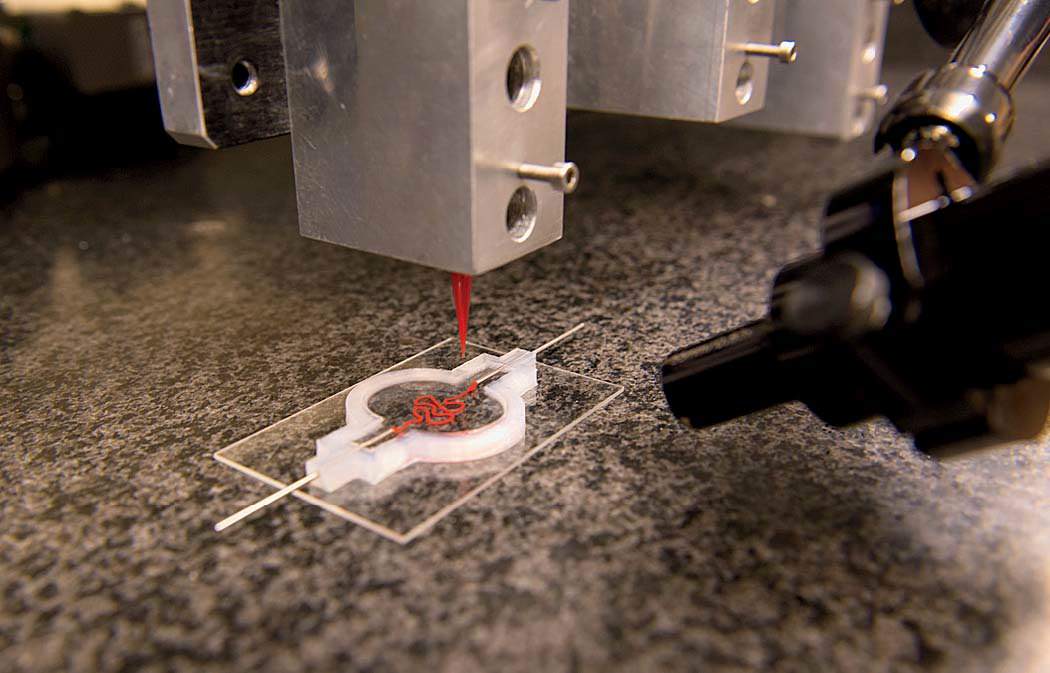
“All right,” says David Kolesky, Ph.D. ’16. “The moment of truth.” As many times as he’s done this, there’s still always that pause. Wearing blue latex gloves and a white lab coat, Kolesky is about to see what the morning’s work has yielded. In front of him on a glass slide is a two-inch chip maybe a quarter-inch deep that he and two lab-mates spent the past few hours making: a translucent rectangle filled with a gelatinous mix of proteins and fibers mimicking the body’s extracellular matrix, the molecules that give support to living cells. And snaking through the middle of it, invisible for the moment, two tiny tubular structures, each a hundred microns or so in diameter (about the width of a human hair)—both produced on the massive black 3-D printer standing a few feet away. One of those tubes, the straighter of the two, will be transformed into a working blood vessel. The other will become something called a proximal tubule, a subcomponent of a nephron—the basic structural and functional unit of the kidney.
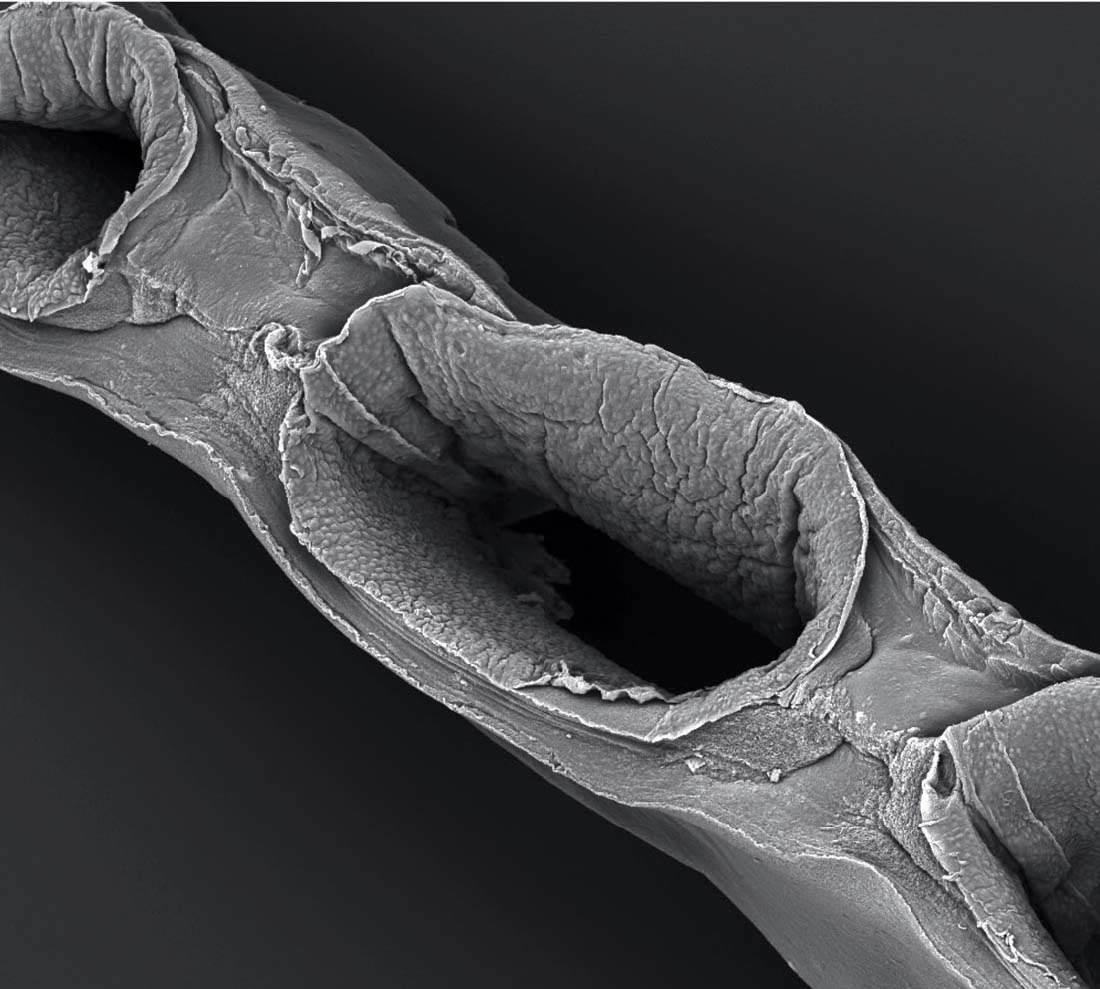
Kolesky is a postdoctoral researcher for materials scientist Jennifer Lewis, the Wyss professor of biologically inspired engineering at the Harvard Paulson School of Engineering and Applied Sciences and a faculty member in the Wyss Institute for Biologically Inspired Engineering. Lewis’s work using 3-D printing technology to fabricate human tissues—“bioprinting,” she calls it—is increasingly important in the emerging field of tissue engineering, a science whose ultimate aspiration is to build three-dimensional tissues and entire organs that can repair or replace a patient’s damaged liver, or heart—or kidney: Lewis and her bioprinting team, whose core members are Kolesky and fellow postdocs Kim Homan and Mark Skylar-Scott, recently constructed a functioning proximal tubule; an October Scientific Reports article laid out the details. Inside the proximal tubule’s convolutions, 65 to 80 percent of nutrients are reabsorbed and transported from the renal filtrate back into the bloodstream. It isn’t a whole kidney yet—it’s not even a whole nephron—but it’s a key subunit, a piece of a piece. It’s a start. “We’re breaking the nephron down into these modular units,” Lewis says, “laying the foundation for the building blocks. We’re testing the structure and function of the modular units with ultimately the goal of trying to assemble a single nephron.”
And from there, eventually, a kidney. That’s the dream.
But many steps lie in between—and many unanswered questions. And so Lewis is careful not to overuse the “big O-word”: organ. Not to over-promise. “We’re still so far away,” she says, from that first fully functioning organ printed in a lab. Along the way, there are very real advances to be made. Bioprinted tissues can offer a more precise platform for modeling disease and screening new drugs. Scientists can see not only whether those drugs work but understand exactly how, in an environment that replicates actual tissue behavior more closely than a two-dimensional layer of cells spread out on a dish, or even animal models. Right now, one in five drugs fails in Phase III clinical trials. That’s a huge failure rate, Lewis says. At that late stage, these potential drugs have been put through multiple layers of preclinical and then clinical testing, with hundreds of millions of dollars and long years of research sunk into their development. “We want to provide a fail-fast step,” she says, “to assess drug toxicity early.” (And not only for the kidney: recently, she collaborated with Wyss Institute colleague Kevin Parker on a report in Nature Materials showing how they used 3-D printing to create a “heart-on-a-chip” model with embedded sensors capable of measuring cardiac tissue responses to drugs and toxic compounds.)
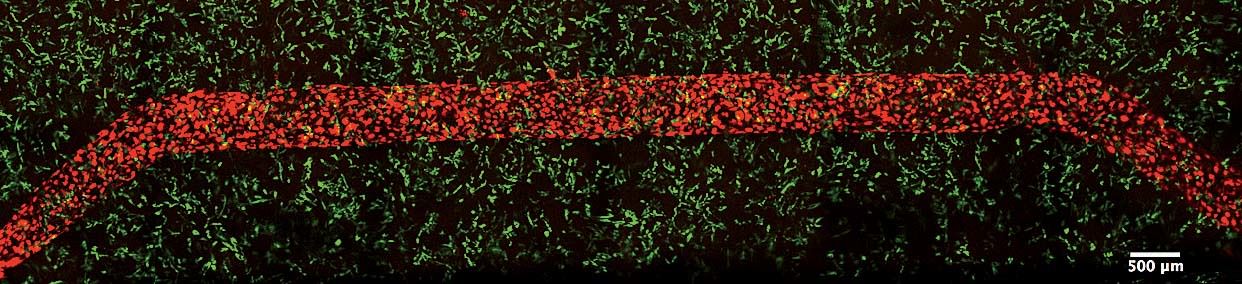
Still, though: the dream is printing organs. And Lewis’s work during the past four years has cleared some difficult hurdles. In 2014, she and several team members, including Kolesky and Homan, published a paper in Advanced Materials describing a way to print tissues made of human cells and vasculature inside an extracellular matrix. For years, this had been a stumbling block in tissue engineering: without blood vessels to carry nutrients in and carry waste out, cells locked in the interior of a printed tissue soon die, even as those along the edges flourish and multiply. The study was a major breakthrough—“our first home run,” Lewis says—and she and her team followed it up last fall by constructing their proximal tubule, a winding, hollow tube that filters nutrients into the bloodstream.
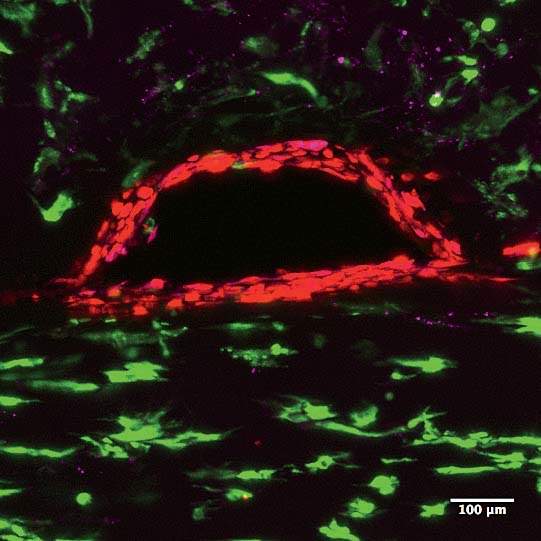
Back in the laboratory, Kolesky, flanked by two undergraduate research assistants with instruments at the ready, pulls two metal pins out from either side of the 3-D-printed chip, releasing a clear liquid from inside the tubes to reveal the two channels they’ve printed: the vascular one and the more serpentine proximal tubule. And suddenly now they’re visible, two perfect thin lines running the length of the chip, white against the milky-gray translucence. Later, Lewis’s team will flow in human epithelial cells, which will stick to the interior walls and grow and proliferate. One channel the scientists will steer toward becoming a proximal tubule, the other toward becoming a blood vessel. Today’s work is part of a broader experiment to see whether Lewis’s team can encourage a proximal tubule to interact with new blood vessels. “In the regions where they’re very, very close, we should see proteins being transported from the kidney side, the tubule, to the vasculature, the blood vessel,” Kolesky says. “That’s exactly what happens in the body. And that’s what we’re trying to make, something that actually behaves like a vascularized proximal tubule.”
The first 3-D printers appeared about 30 years ago. In 1984 an American engineer named Charles Hull patented what he called a stereolithography apparatus, a machine that could make solid objects by “printing” them out in thin layers laid one on top of another. The “ink” in Hull’s machine was an acrylic liquid that turned hard when exposed to ultraviolet light. The earliest applications for this technology were industrial: computer-designed prototypes for complex parts in airplanes and cars and other vehicles, customized equipment components that could be made quickly and cheaply and on demand. Inventors could test out their creations without much upfront manufacturing cost.
These days, 3-D printers are everywhere—in stores and offices and homes (the first desktop 3-D printer went on sale in 2009)—and they make all kinds of things: earrings and necklaces, eyeglass frames, fitted insoles and midsoles for running shoes, smartphone cases, action figures (and not just of superheroes—customers can upload photos and have figures made of themselves), souvenir replicas of the Eiffel Tower and the Statue of Liberty and Michelangelo’s David. Car bodies and concrete walls for houses can be built with 3-D printers, which can print plastics, metals, ceramics, wax, and even food.
The technology has long been used in medical settings too, to custom-build stents, prosthetics, hearing aid shells, acrylic dental crowns, orthodontic braces, and detailed surgical models on which physicians can practice. In 2014, doctors in the Netherlands saved a woman’s life by replacing most of her skull with a 3-D-printed one made from plastic.
The first person to print out biological materials was Thomas Boland, a biomedical engineer who in 2003 filled the ink cartridge of a Lexmark inkjet printer with collagen, and later E. coli cells, and then mammalian cells from rats and hamsters. Ninety percent of the cells remained alive after printing.
The chase to build a fully functioning human organ began in earnest in 2007, when Gabor Forgacs, a biological physicist at the University of Missouri, printed spherical particles containing several different types of chicken heart cells onto large sheets of gel-laden paper. After they were printed, the cells began to coalesce into a single structure, like droplets of water coming together. And then they began to beat.
Lewis exudes the coiled energy of an athlete at rest. With her close-cropped hair, wire-rimmed glasses, and button-down shirts rolled to the elbow, she is amiably intense. Even when she’s seated at her desk, you feel like she might be on the balls of her feet.
She started out not in biology, but materials science. Raised just outside Chicago, she went to college at the University of Illinois at Urbana-Champaign and as a freshman joined the ceramic-engineering program. Later, at MIT, she got a doctorate in ceramic science. Ceramics are inorganic and nonmetallic, and they possess unusual diversity and breadth of behaviors: they can form electronic components, ultrasound transducers, high-temperature insulation—like the tiles that coat the space shuttle—as well as window glass and pottery.
After MIT, Lewis returned to Urbana-Champaign in 1990, where she established her own research group and, working with materials scientists Scott White and Nancy Sottos, developed a self-healing plastic that could repair itself over and over when damaged, not unlike the way human skin does. The researchers’ approach was to embed a three-dimensional network of microchannels throughout the material—like veins and arteries—so that when a crack formed, a liquid healing agent inside this nexus would wick toward the damage and polymerize. Essentially, it would form a “clot.”
To create those microchannels, Lewis and her colleagues at Illinois used a 3-D printable ink that would melt when it was heated. But in 2011, Lewis began formulating a new ink, one that at room temperature had roughly the consistency of toothpaste but turned to liquid when cooled to just above freezing. Using this ink, she could print hydrogels (supportive gelatinous substances) filled with elaborate vascular networks, then lower the temperature until the ink liquefied and remove it, leaving a system of open channels. Lewis coined the term “fugitive ink” to describe her innovation.
It wasn’t hard to see that this method, inspired by biology, might have biological uses.
In 2013, Lewis came to Harvard, with bioprinting on her mind. “We moved here primarily to launch this entirely new effort,” she says, “building on our expertise in materials, in design of materials, in 3-D printing of materials.” Before arriving in Cambridge, she had never worked with cells directly in her own lab. Several graduate students made the cross-country move with her; among them was Kolesky, who scrapped his original doctoral focus in materials science to do a thesis on bioprinting of vascularized human tissues. “We’re materials scientists gone rogue,” he jokes.
Bioprinting isn’t the lab’s only focus: her team is also involved in soft electronics, robotics, and composites, all relying on 3-D printing. The two dozen graduate students, undergraduates, and postdocs have printed wearable soft sensors, antennas made from conductive silver nanoparticles, and rechargeable microbatteries on a millimeter scale to power small wireless sensors and medical devices. In 2016 her lab, led by Skylar-Scott, pioneered a technique for printing 3-D metal structures in midair—without the supportive substrate previously required—and, collaborating with Wyss Institute researcher Robert Wood, built the first-ever completely soft, autonomous robot, in the shape of an octopus. Nicknamed “octobot,” the small and surprisingly charismatic pneumatic device was made from silicone gel and powered by a chemical reaction that pushed gas through the chambers in its rubbery legs. The work was published in Nature. That same year, Lewis and her team used a hydrogel composed with cellulose fibers to create a series of “four-dimensionally printed” flowerlike structures that change shape when they’re immersed in water—twisting, ruffling, and bending the way plant leaves do in response to environmental stimuli.
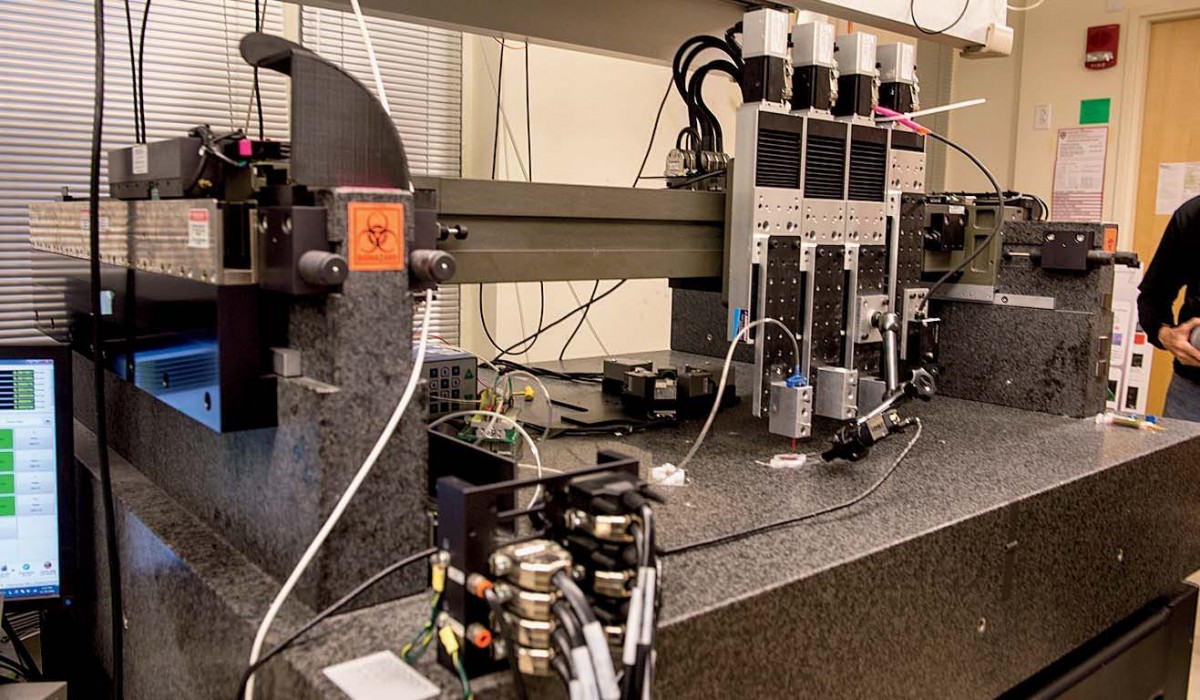
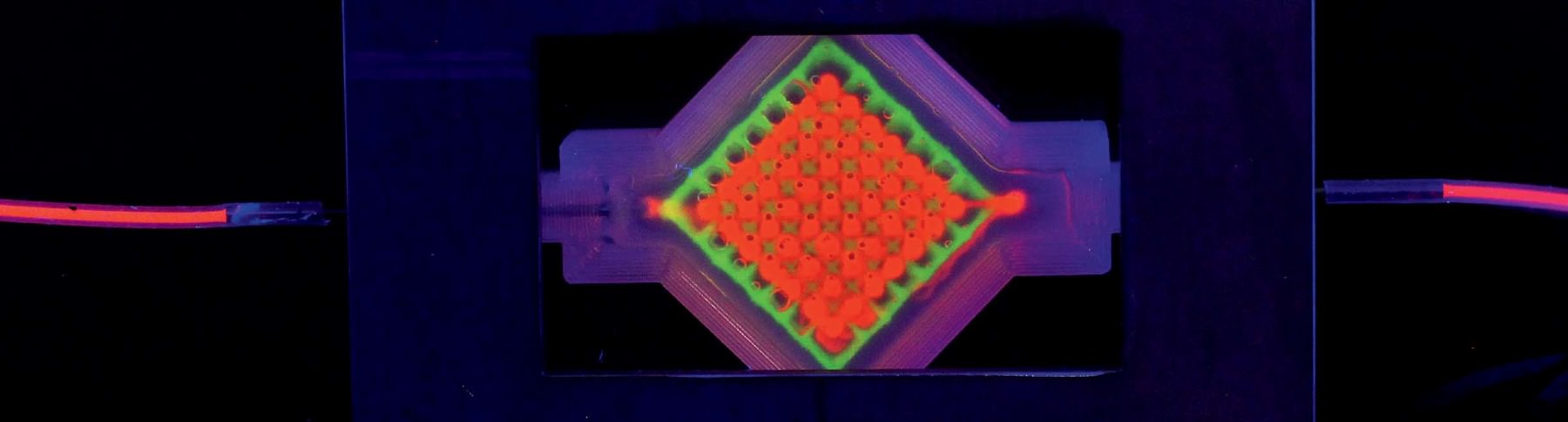
Increasingly, though, the central focus of Lewis’s research is tissue engineering. (In October, the Wyss Institute launched a cross-cutting initiative aimed at 3-D organ engineering, co-led by Lewis and Christopher Chen, who runs Boston University’s tissue microfabrication laboratory.) The 3-D printer Lewis’s team uses for bioprinting, one of four printers in her lab, is colossal. It sits on supports at the back of the busy maze of her lab: a black enormity with a granite base that weighs a ton and a half—the weight provides stability for the printer’s fine-scale work. Above sit the ink dispensers, mounted on four rectangular steel plates that can move independently, allowing for multiple inks to be sent through the tiny nozzles at the bottom of each dispenser. A tangle of tubes and wires connect the dispensers to the printer, whose motion is controlled by a computer that sits just off to the left.
In February 2014, almost a year to the day after her arrival at Harvard, Lewis and her team published the Advanced Materials paper describing how fugitive ink could be used to print channels for tissue vasculature. “That was the big advance,” Kolesky says. “That made everything else possible—this sacrificial material that templates out a channel, and then just goes away,” leaving an opening for the team to inject with cells to line its walls. Skylar-Scott, who came to Lewis’s lab with an undergraduate degree in electrical engineering and a Ph.D. in biomedical engineering, offers a bit of context: every cell in the human body is only about 200 microns away from the nearest blood vessel, he says. Even the thinnest tissues, like skin, are highly vascularized, covered in nerve endings and capillaries.
Homan explains further: “The reason this ink was so critical is because other fugitive materials require high temperatures or other crazy treatments for removal that would kill the cells nearby.” Lewis’s requires only 10 minutes in the fridge.
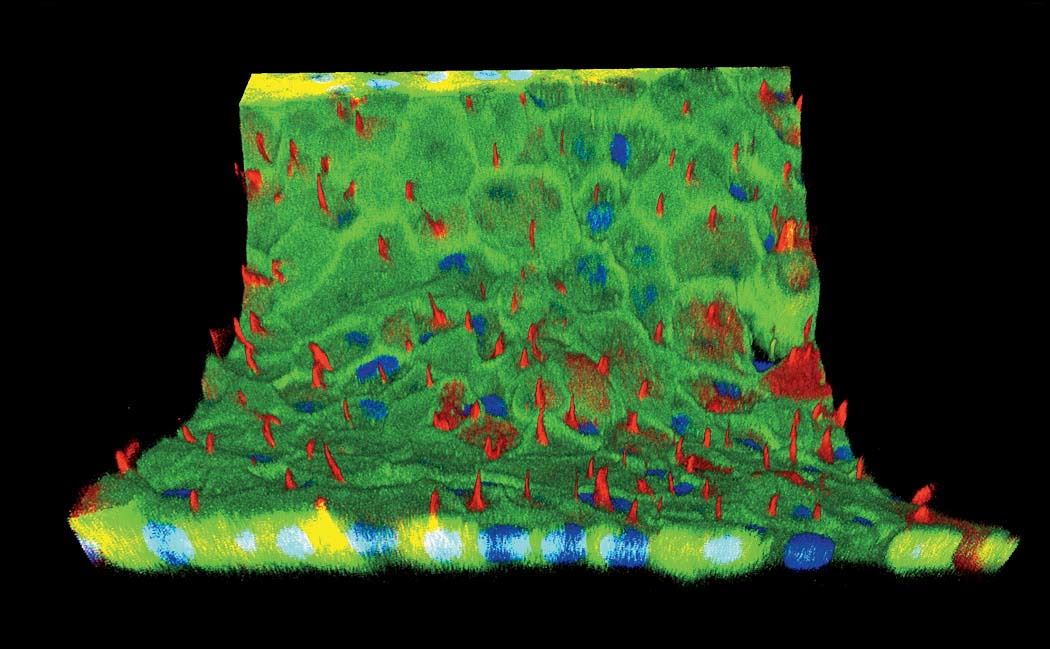
Then last year in Proceedings of the National Academy of Sciences, Lewis’s team demonstrated the effectiveness of their bioprinting approach by creating a multi-layer tissue, more than a centimeter thick (most other printed tissues have been less than a millimeter thick), combining vasculature with printed human stem cells that ultimately become a bone-like tissue. “We differentiated these cells down an osteogenic pathway, so they started laying down calcium phosphate, the primary mineral in bone, within the tissue,” Lewis says. The tissue survived for more than six weeks. Inside the chips, cells reorganize, they grow, they take on a life of their own and remodel their environment. “We went up to about a centimeter cubed thickness,” Kolesky says. “That’s about 10 times thicker than what people have been doing—and we’re not sure what the limit is, or if there is a limit.”
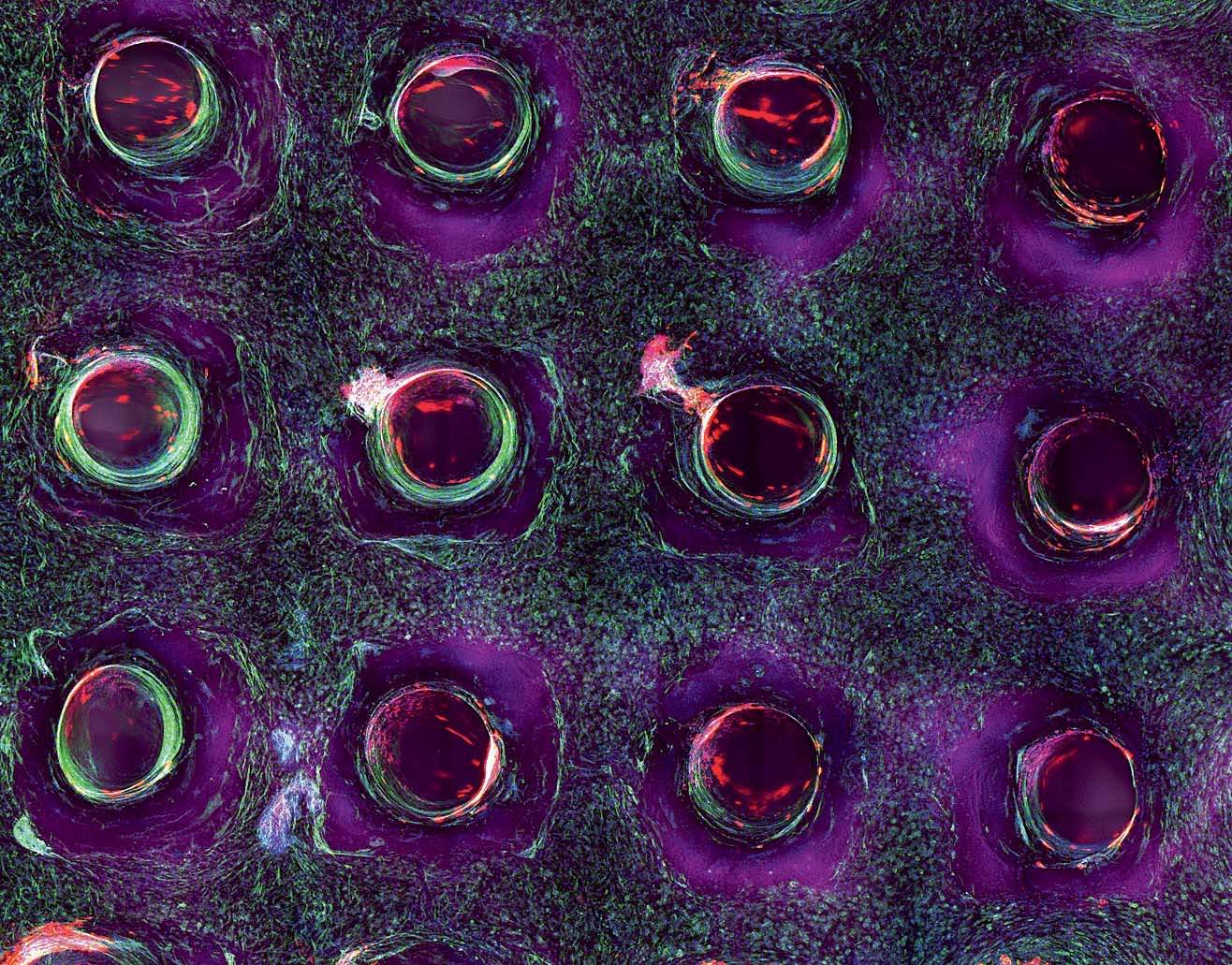
Structure matters. The difference between a single layer of cells on a two-dimensional dish and full-fledged, fully functioning tissue, even if it’s just a centimeter thick, is tremendous. “They take on a significantly different morphology,” Homan says. “They behave in such different ways.” In the paper they published last fall on printing proximal tubules, Lewis and her team showed that the cells in their 3-D system grew taller than in a monolayer of kidney cells on a dish. Hair-like structures called microvilli, which stick out into the center of the tube and absorb nutrients and other media flowing past, were much longer and more robust in the 3-D cells, and the extra-long primary cilia, sometimes missing from cells raised on a dish, were clearly visible.
Until a few years ago, the mainstay of biological research was a cell culture on a dish. As Skylar-Scott explains: “That’s where all these pathways were found, where a lot of drugs were tested, and starting in the early 2000s, around the same time as the Genome Project, a few key organisms”—the C. elegans worm, the fruit fly, the zebrafish, the mouse, the chimpanzee—“were selected for close study so that we could use them as three-dimensional models to gain an understanding of biology. So essentially you have these half-dozen studied-to-death organisms because biologists realized that 2-D cell culture was just not telling them how things really work. We don’t exist as a single cell type on a flat surface. Cell-to-cell signaling isn’t just a byproduct of biology; it is biology. And it doesn’t exist in a dish.”
Lewis and her team began working on the kidney a little more than two years ago, after they decided to take the “moonshot” toward the O-word. Having already shown that they could print tissue and keep it alive for months at a time by threading blood vessels through it, Lewis wanted to focus on tissue that would not merely be “demonstrative,” she says, but also functional. As Homan puts it, “Functionality is the point.”
Kidneys are badly needed. Of the roughly 120,000 Americans on the national organ-donor waiting list, more than 100,000 are waiting for kidneys. Every year 4,500 people die without transplants. As the population ages and obesity increases, there are ever more patients with diabetes or on dialysis, or with acute kidney failure. (Lewis’s kidney research is funded in part by the Swiss pharmaceutical company Roche, and Annie Moisan, a Roche scientist with whom Lewis began collaborating two years ago, coauthored the Scientific Reports study.) In a 2015 talk at Dartmouth, Lewis described some of the emails she and her team had begun receiving from ordinary strangers out in the world, people she didn’t know who happened to hear about her work. The talk’s moderator, Dartmouth computer scientist Dan Rockmore, asked her whether in the course of fabricating living tissue, she had ever heard from people accusing her of playing God. No, she said. “I get the more heartbreaking letters, actually: ‘My daughter or my son—’ and ‘When is this going to be available?’ And we’re still such a long way from something that can have an impact in the clinic.…But those are the emails that are difficult. Because you want to be able to help people. And they motivate you.”
Shaped like a long, curving filament, the proximal tubule is a complicated piece of renal architecture. Circulating drugs and their metabolites can accumulate in high concentrations there, and when the kidney is damaged, proximal tubules are most often the site. “This is a foundational building block,” Lewis says. But it’s not the only one. There are other important subunits to build: the glomerus, where the nephron’s filtering process begins, and the Bowman’s capsule that surrounds it, and other, smaller components. “Right now we’re trying to construct a single nephron,” Lewis says. In a human kidney, there are a million of them. “But once you have the nephron, then it gets interesting. Once you can say, ‘We can construct a nephron and it’s fully functional’—and to be clear, we’re not there yet—then it’s like saying, ‘We have a transistor.’” She searches through the files on her computer and brings up an image of a graph with a line sloping sharply upward. “Here’s the first transistor, in 1947,” she says, pointing. That was the year it was invented, at Bell Labs. “It took 25 years just to get to this, a single solid-state transistor,” the basis of all computer chips. “And from there, it took very little time for this rapid upward trajectory, so that now you have computer chips made of billions of transistors.…Right now, we’re still here at the bottom. We have a part of one nephron. But we’re getting closer to having a complete one—and once we’ve got one, the key question is, will it follow this same trajectory? I don’t know. But this is illustrative. It tells you what might be possible.”

As the team moves from the proximal tubule to other components of the kidney, there are basic science questions to be asked and answered about how, precisely, kidneys function. “We’ve never before been able to put two tubules close together to see vectorial transport—the mechanism by which nutrients are sent back into the bloodstream and everything else is sent to the urine,” says Homan. “There are a ton of questions.” A central one relates to the problem of scaling up: in other words, figuring out just how much of the kidney, as it exists in the body, must be replicated in order for a replacement kidney to work. There are about 150 billion cells in an adult kidney, and the organ’s microvasculature, Skylar-Scott says, “probably stretches from here to San Francisco”—do scientists have to print every one? That would take days and days of continuous printing, and cells can survive for only a couple of hours outside their nourishing media. “We set a timer as soon as we lift the cells off the plate” to print them, he says. “You watch the timer and you work fast. It takes more than one person to do this. That’s why it’s always been a team effort amongst us.” Adds Homan: “You really don’t want to bioprint alone.” Meanwhile, printing cells too quickly can damage them.
A similar question of scaling confronts the formation of vasculature. “The smallest blood vessels that we print are usually about 400 microns in diameter,” Kolesky says. The very thinnest ones they’ve printed are about 75 microns. “But the smallest blood vessels in the body are 5 to 10 microns. If we have to build a tissue capillary by capillary, it’s going to take forever.…Can we print the larger-scale blood vessels and have the smaller ones assemble on their own?” In the body, that process is called angiogenesis. Lewis likes to describe it using an analogy: “We lay down the highways,” she says, “and then let the cells create the smaller roads and driveways.” Skylar-Scott explains, “If you take your arteries and arterioles and veins and stretch them all out, they probably wouldn’t reach the end of this room. But you exponentially grow in length when you get down to the scale of capillaries. That’s when you’re talking about hundreds of thousands of miles of those tiniest blood vessels throughout your body.” In creating vasculature, the aim is to encourage biology to build as much of the tissue as possible.
The same principle goes for entire organs. “We want to elucidate what part of the organ we have to print, and what part we can leave to biology,” Lewis says. “The stem cells themselves are going to interact with the matrix around them and lay down their own matrix. They’re going to grow and proliferate, and differentiate into different cell lineages.…We really don’t want to construct an entire organ, cell by cell, capillary by capillary. That’s a daunting challenge. But what we don’t know yet, what we haven’t yet teased out, is how far down the path we have to go before biology will do the rest, before you can implant something in a patient that the body will accept. What are the right cell types to print and with what spatial precision and what density? How close to that billion cells per milliliter do we need to get to recapitulate function?” When is a kidney a kidney?
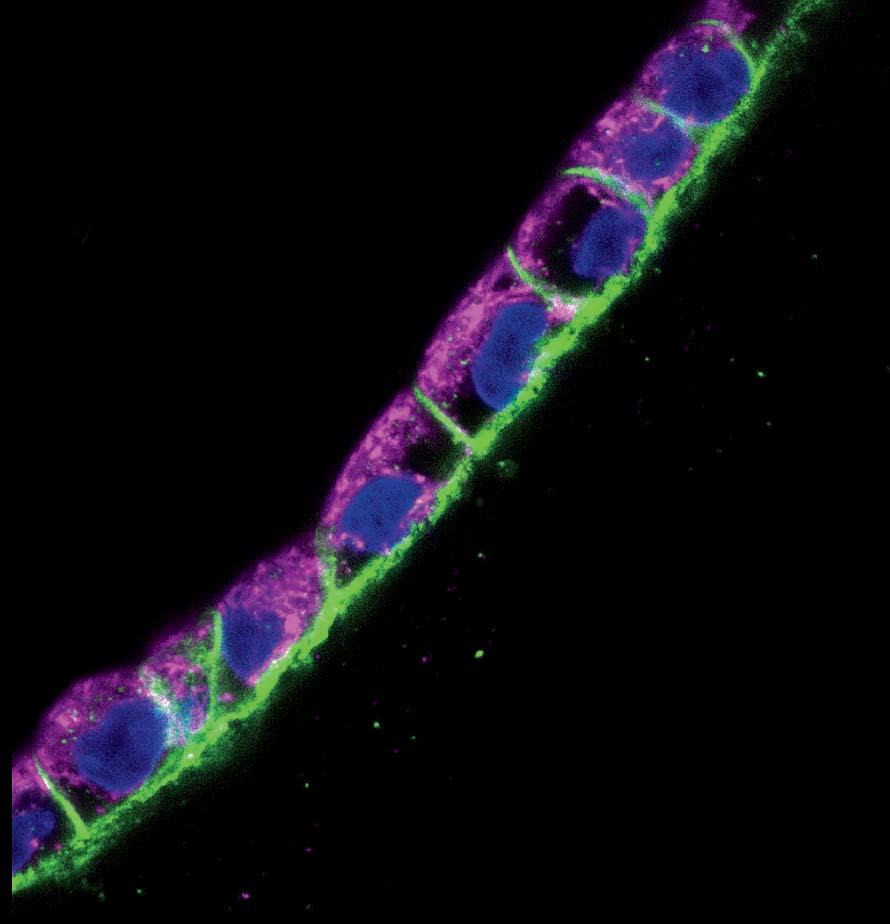
In a darkened, closet-sized room off the lab, Homan sits gazing into a monitor attached to a confocal-laser scanning microscope. On a dish under the lens are cross-sectional slivers of proximal tubules, which, after living and growing inside the chip for four weeks, have been sliced up and stained with antibodies that attach to specific proteins. “We chop it up to see what it’s been doing, what properties it’s exhibited, and how it’s behaving at a protein-expression level,” she says. She’s looking to see if the cells in the proximal tubule are functioning the way they would in the human body. It’s slow, careful work. Not every sample yields an answer, and it can take hours for Homan to find what she’s looking for. When she activates the microscope’s laser, the antibodies light up if the protein is present: blue, green, red, purple. “I look for a lot of things: cell height, how tightly packed they are, any sign of contamination, how well the cells stain, if the stain is supposed to be on one side of the cell versus the other side.…We also look at their uptake: we might send a protein into the tube while it’s still in the incubator and then look at the cells’ ability to uptake that protein.”
Today she’s scanning for six or eight different proteins. “We’re looking for that,” she says, bringing up a microscope image taken from an earlier sample. It’s gorgeous, almost like a photograph captured from space: a chain of tightly packed cells curving up and to the right, blue-stained nuclei encased in walls of green and purple, each color representing a different protein. “It’s nice, right?” Today, though, she’s still looking. Hours later, she’s refilled her water bottle and eaten a banana and a cup of yogurt. She returns to the microscope. “I’m going to keep at it,” she says.